5C6G
 
 | |
5GWE
 
 | | cytochrome P450 CREJ | | Descriptor: | (4-methylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Dong, S, liu, X, Wang, X, Feng, Y. | | Deposit date: | 2016-09-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8KCJ
 
 | | De novo design protein -N7 | | Descriptor: | De novo design protein -N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo design protein -N7
To Be Published
|
|
8KBB
 
 | | Structure of apo-CmTad1 | | Descriptor: | Thoeris anti-defense 1, ZINC ION | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of apo-CmTad1
To Be Published
|
|
8SBG
 
 | |
6MMC
 
 | |
8KA6
 
 | | De novo design protein -NA7 | | Descriptor: | De novo design protein -NA7 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De novo design protein -NA7
To Be Published
|
|
5TX5
 
 | | Rip1 Kinase ( flag 1-294, C34A, C127A, C233A, C240A) with GSK772 | | Descriptor: | 3-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1H-1,2,4-triazole-5-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P, Thrope, J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases.
J. Med. Chem., 60, 2017
|
|
5TXI
 
 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-16 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5C92
 
 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, in complex with copper | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (I) ION, ... | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
8KB9
 
 | |
5TOR
 
 | | Crystal structure of AAT D222T mutant | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
8KC5
 
 | | De novo design protein -T09 | | Descriptor: | De novo design protein -T09 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo design protein -T09
To Be Published
|
|
6MV9
 
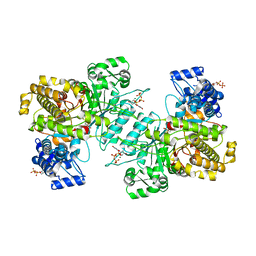 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
7P2S
 
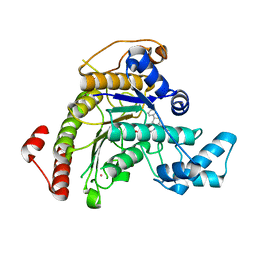 | | Crystal structure of Schistosoma mansoni HDAC8 in complex with a tricyclic thieno[3,2-b]indole capped hydroxamate-based inhibitor, chlorine derivative | | Descriptor: | 5-[[(2R)-7-fluoranyl-2-phenyl-2,3-dihydrothieno[3,2-b]indol-4-yl]methyl]-N-oxidanyl-thiophene-2-carboxamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Saccoccia, F, Gemma, S, Campiani, G, Ruberti, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Schistosoma mansoni histone deacetylase 8 reveal a novel binding site for allosteric inhibitors.
J.Biol.Chem., 298, 2022
|
|
7VDY
 
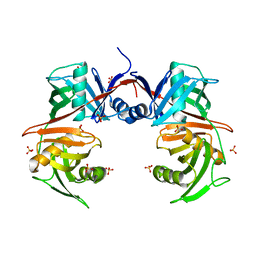 | | Crystal structure of O-ureidoserine racemase | | Descriptor: | O-ureido-serine racemase, SULFATE ION | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of O-ureidoserine racemase found in the d-cycloserine biosynthetic pathway.
Proteins, 90, 2022
|
|
7MBH
 
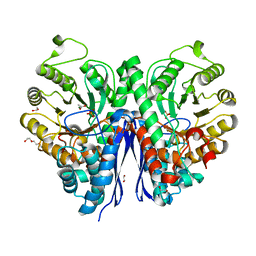 | | Structure of Human Enolase 2 in complex with phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Gamma-enolase, ... | | Authors: | Leonard, P.G, Hicks, K.G, Rutter, J. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-metabolite interactomics of carbohydrate metabolism reveal regulation of lactate dehydrogenase.
Science, 379, 2023
|
|
7OOO
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG containing an LNA-Amide-LNA modification | | Descriptor: | DNA (5'-D(*CP*TP*(05A)P*TP*CP*TP*TP*TP*G)-3'), MAGNESIUM ION, RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-05-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
7OOS
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG | | Descriptor: | DNA (5'-D(*CP*TP*(05K)P*TP*CP*TP*TP*TP*G)-3'), RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), STRONTIUM ION | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-05-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
7OZZ
 
 | | The crystal structure of a DNA:RNA hybrid duplex sequence CTTTTCTTTG with LNA-amide modification | | Descriptor: | DNA (5'-D(*CP*TP*(05H)P*TP*CP*TP*TP*TP*G)-3'), POTASSIUM ION, RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3') | | Authors: | Thorpe, C, Hardwick, J, McDonough, M.A, Hall, J.P, Baker, Y.R, El-Sagheer, A.H, Brown, T. | | Deposit date: | 2021-06-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An LNA-amide modification that enhances the cell uptake and activity of phosphorothioate exon-skipping oligonucleotides.
Nat Commun, 13, 2022
|
|
5U10
 
 | |
7P43
 
 | | Structure of CgGBE in complex with maltotriose | | Descriptor: | 1,4-alpha-glucan-branching enzyme, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ballut, L, Conchou, L, Violot, S, Galisson, F, Aghajari, N. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Candida glabrata glycogen branching enzyme structure reveals unique features of branching enzymes of the Saccharomycetaceae phylum.
Glycobiology, 32, 2022
|
|
5TPT
 
 | |
6MVS
 
 | | Structure of a bacterial ALDH16 complexed with NAD | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tanner, J.J, Liu, L. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J. Mol. Biol., 431, 2019
|
|
8KDK
 
 | |
