8GYM
 
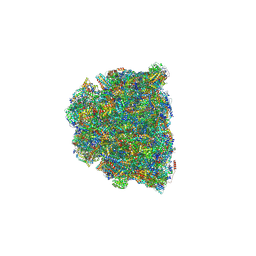 | | Cryo-EM structure of Tetrahymena thermophila respiratory mega-complex MC IV2+(I+III2+II)2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
8GZU
 
 | | Cryo-EM structure of Tetrahymena thermophila respiratory Megacomplex MC (IV2+I+III2+II)2 | | Descriptor: | 2 iron, 2 sulfur cluster-binding protein, 2-oxoglutarate/malate carrier protein, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-27 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
1R7S
 
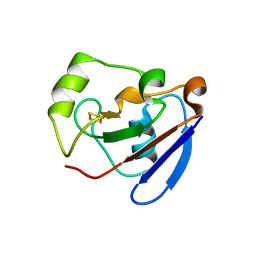 | | PUTIDAREDOXIN (Fe2S2 ferredoxin), C73G mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Smith, N, Mayhew, M, Kelly, H, Robinson, H, Heroux, A, Holden, M.J, Gallagher, D.T. | | Deposit date: | 2003-10-22 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of C73G putidaredoxin from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8HKD
 
 | |
3LB8
 
 | |
2IAG
 
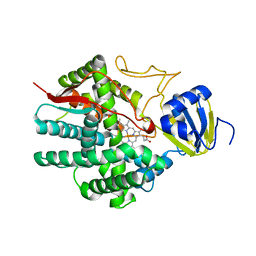 | | Crystal structure of human prostacyclin synthase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Prostacyclin synthase, SODIUM ION | | Authors: | Chiang, C.-W, Yeh, H.-C, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2006-09-08 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human Prostacyclin Synthase
J.Mol.Biol., 364, 2006
|
|
2WLB
 
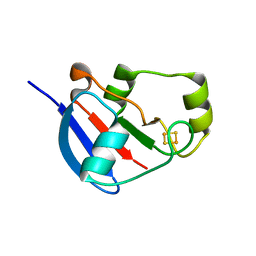 | | Adrenodoxin-like ferredoxin Etp1fd(516-618) of Schizosaccharomyces pombe mitochondria | | Descriptor: | ELECTRON TRANSFER PROTEIN 1, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Mueller, J.J, Hannemann, F, Schiffler, B, Bernhardt, R, Heinemann, U. | | Deposit date: | 2009-06-23 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Characterization of the Adrenodoxin-Like Domain of the Electron-Transfer Protein Etp1 from Schizosaccharomyces Pombe.
J.Inorg.Biochem., 105, 2011
|
|
1AYF
 
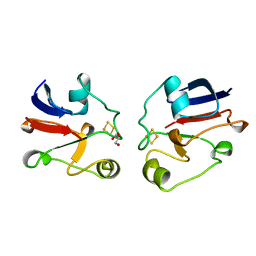 | | BOVINE ADRENODOXIN (OXIDIZED) | | Descriptor: | ADRENODOXIN, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL | | Authors: | Mueller, A, Mueller, J.J, Heinemann, U. | | Deposit date: | 1997-11-03 | | Release date: | 1998-12-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New aspects of electron transfer revealed by the crystal structure of a truncated bovine adrenodoxin, Adx(4-108).
Structure, 6, 1998
|
|
1XLQ
 
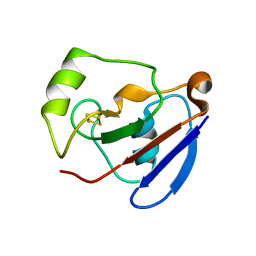 | |
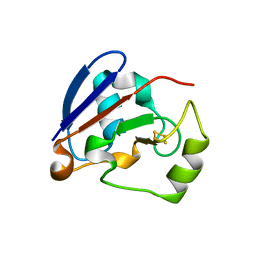
1XLN
 
 | |
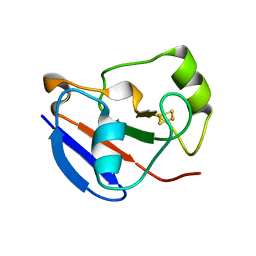
1XLP
 
 | |
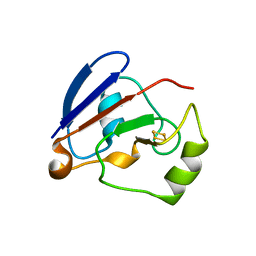
1XLO
 
 | |
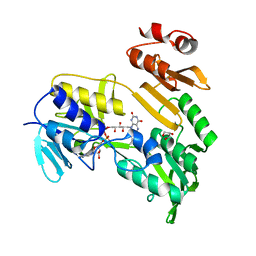
3FG2
 
 | |
4UVR
 
 | | Binding mode, selectivity and potency of N-indolyl-oxopyridinyl-4- amino-propanyl-based inhibitors targeting Trypanosoma cruzi CYP51 | | Descriptor: | Nalpha-{4-[4-(5-chloro-2-methylphenyl)piperazin-1-yl]-2-fluorobenzoyl}-N-pyridin-4-yl-D-tryptophanamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-DEMETHYLASE, ... | | Authors: | Vieira, D.F, Choi, J.Y, Calvet, C.M, Gut, J, Kellar, D, Siqueira-Neto, J.L, Johnston, J.B, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Binding Mode and Potency of N-Indolyl-Oxopyridinyl-4-Amino-Propanyl-Based Inhibitors Targeting Trypanosoma Cruzi Cyp51
J.Med.Chem., 57, 2014
|
|
7RTQ
 
 | | Sterol 14alpha demethylase (CYP51) from Naegleria fowleri in complex with an inhibitor R)-N-(1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | N-[(1R)-2-(1H-imidazol-1-yl)-1-(3,4',5-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, Protein CYP51 | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Wawrzak, Z. | | Deposit date: | 2021-08-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Relaxed Substrate Requirements of Sterol 14 alpha-Demethylase from Naegleria fowleri Are Accompanied by Resistance to Inhibition.
J.Med.Chem., 64, 2021
|
|
6E8Q
 
 | | S. CEREVISIAE CYP51 COMPLEXED WITH Posaconazole | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Lanosterol 14-alpha demethylase, PENTAETHYLENE GLYCOL, ... | | Authors: | Tyndall, J.D, Keniya, M.V, Sabherwal, M, Monk, B.C. | | Deposit date: | 2018-07-30 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Azole Resistance Reduces Susceptibility to the Tetrazole Antifungal VT-1161.
Antimicrob. Agents Chemother., 63, 2019
|
|
8Q7E
 
 | |
8Q7H
 
 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SBI
 
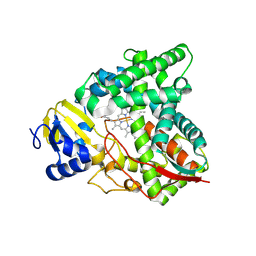 | |
4UHL
 
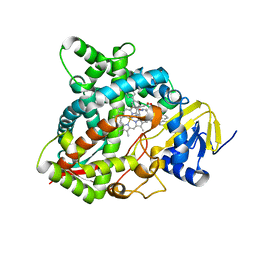 | | HUMAN STEROL 14-ALPHA DEMETHYLASE (CYP51) IN COMPLEX WITH VFV IN P1 SPACE GROUP | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, I Lepesheva, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human Sterol 14Alpha-Demethylase (Cyp51) as a Target for Anticancer Chemotherapy: Towards Structure-Aided Drug Design.
J.Lipid Res., 57, 2016
|
|
4UHI
 
 | | HUMAN STEROL 14-ALPHA DEMETHYLASE (CYP51) IN COMPLEX WITH VFV IN C121 SPACE GROUP | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, I Lepesheva, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Human Sterol 14Alpha-Demethylase (Cyp51) as a Target for Anticancer Chemotherapy: Towards Structure-Aided Drug Design.
J.Lipid Res., 57, 2016
|
|
6RQ5
 
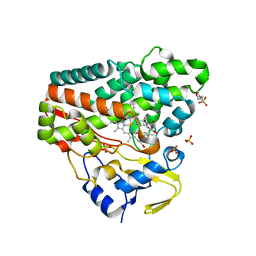 | | CYP121 in complex with 3,5-dimethyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(3,5-dimethyl-4-oxidanyl-phenyl)methyl]-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
7RY8
 
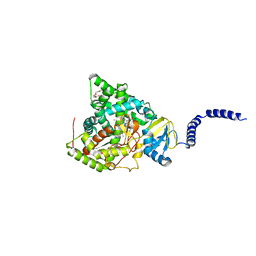 | | S. CEREVISIAE CYP51 Y140H mutant COMPLEXED WITH Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PENTAETHYLENE GLYCOL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
7QQ7
 
 | |
4CK8
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH (R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl 4-(4-(3,4- dichlorophenyl)piperazin-1-yl)phenylcarbamate (LFD) | | Descriptor: | (1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl {4-[4-(3,4-dichlorophenyl)piperazin-1-yl]phenyl}carbamate, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Friggeri, L, Wawrzak, Z, Tortorella, S, Lepesheva, G.I. | | Deposit date: | 2013-12-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma Cruzi Sterol 14Alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition.
J.Med.Chem., 57, 2014
|
|
