1H17
 
 | |
7YJM
 
 | | Cryo-EM structure of the monomeric atSPT-ORM1 complex | | Descriptor: | Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJO
 
 | | Cryo-EM structure of the monomeric atSPT-ORM1 (LCB2a-deltaN5) complex | | Descriptor: | Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJK
 
 | | Cryo-EM structure of the dimeric atSPT-ORM1 complex | | Descriptor: | Long chain base biosynthesis protein 1, Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
5ONY
 
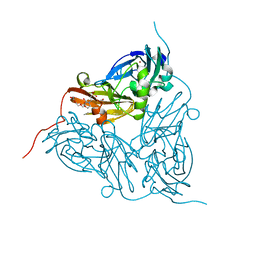 | | As-isolated resting state copper nitrite reductase from Achromobacter xylosoxidans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
7VF3
 
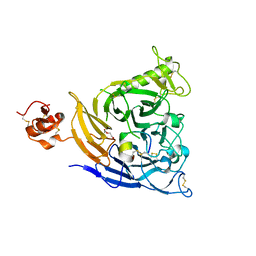 | | Plexin B1 extracellular fragment in complex with lasso-grafted PB1m7 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Plexin-B1, ... | | Authors: | Sugano, N.N, Hirata, K, Yamashita, K, Yamamoto, M, Arimori, T, Takagi, J. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | De novo Fc-based receptor dimerizers differentially modulate PlexinB1 function.
Structure, 30, 2022
|
|
5ONX
 
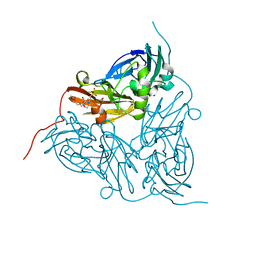 | | Resting state copper nitrite reductase determined by serial femtosecond rotation crystallography | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
7FQJ
 
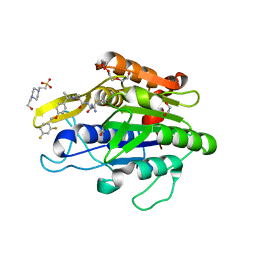 | | Crystal Structure of human Legumain in complex with (2S)-N-[(1S)-3-amino-1-cyano-3-oxopropyl]-1-[1-[4-[(2,4-difluorophenyl)methoxy]phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
1H8E
 
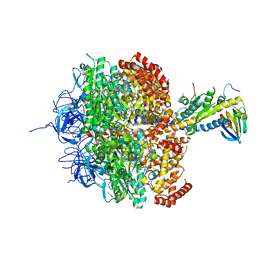 | | (ADP.AlF4)2(ADP.SO4) bovine F1-ATPase (all three catalytic sites occupied) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, GLYCEROL, ... | | Authors: | Menz, R.I, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2001-02-02 | | Release date: | 2001-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase with Nucleotide Bound to All Three Catalytic Sites: Implications for the Mechanism of Rotary Catalysis
Cell(Cambridge,Mass.), 106, 2001
|
|
5RC0
 
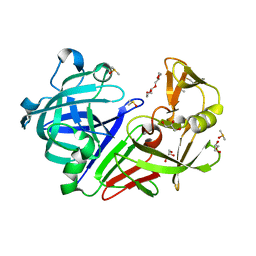 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library E06a | | Descriptor: | (3-endo)-8-benzyl-8-azabicyclo[3.2.1]octan-3-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCD
 
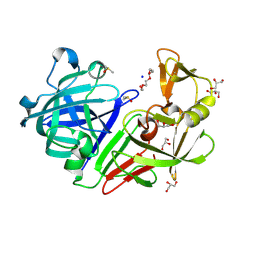 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library H03a | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCT
 
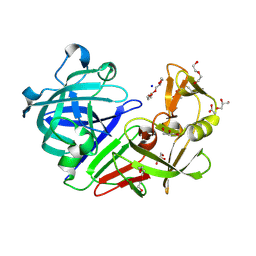 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 15 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7FZT
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with rac-(1R,2S)-2-[(3,4-dichlorobenzoyl)amino]cyclohexane-1-carboxylic acid | | Descriptor: | (1R,2S)-2-(3,4-dichlorobenzamido)cyclohexane-1-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a human FABP4 binding site mutated to that of FABP5 complex
To be published
|
|
7XS6
 
 | | structure of a membrane-integrated glycosyltransferase with inhibitor | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase 1, ... | | Authors: | Wu, Y.N, Zhang, M, Yang, Y.Z, Ding, X.Y, Liu, X.T, Zhang, M.J, Yu, H.J. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | structure of a membrane-integrated glycosyltransferase with inhibitor
To Be Published
|
|
7XS7
 
 | | structure of a membrane-integrated glycosyltransferase | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, Chitin synthase 1, DODECANE, ... | | Authors: | Wu, Y.N, Zhang, M, Yang, Y.Z, Ding, X.Y, Liu, X.T, Zhang, M.J, Yu, H.J. | | Deposit date: | 2022-05-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | structure of a membrane-integrated glycosyltransferase with inhibitor
To Be Published
|
|
2PKT
 
 | | Crystal structure of the human CLP-36 (PDLIM1) bound to the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Uppenberg, J, Gileadi, C, Elkins, J, Bray, J, Burgess-Brown, N, Salah, E, Gileadi, O, Bunkoczi, G, Ugochukwu, E, Umeano, C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-18 | | Release date: | 2007-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms
Protein Sci., 19, 2010
|
|
2PGZ
 
 | | Crystal structure of Cocaine bound to an ACh-Binding Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COCAINE, Soluble acetylcholine receptor, ... | | Authors: | Hansen, S.B, Taylor, P. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Galanthamine and non-competitive inhibitor binding to ACh-binding protein: evidence for a binding site on non-alpha-subunit interfaces of heteromeric neuronal nicotinic receptors.
J.Mol.Biol., 369, 2007
|
|
2PYF
 
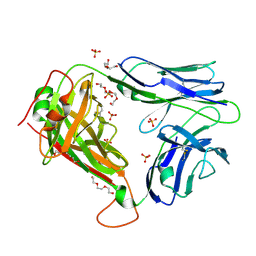 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry Unbound TCR Clone 5-1 | | Descriptor: | SULFATE ION, T-Cell Receptor, Alpha Chain, ... | | Authors: | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | Deposit date: | 2007-05-16 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
1A2I
 
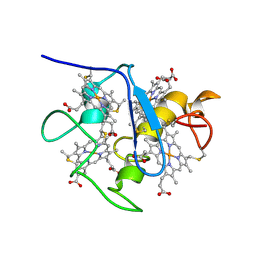 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Kastrau, D.H.K, Costa, H.S, Legall, J, Turner, D.L, Santos, H, Xavier, A.V. | | Deposit date: | 1998-01-05 | | Release date: | 1998-07-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Desulfovibrio vulgaris (Hildenborough) ferrocytochrome c3: structural basis for functional cooperativity.
J.Mol.Biol., 281, 1998
|
|
7WAS
 
 | |
7E0W
 
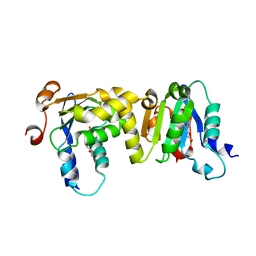 | | Crystal Structure of BCH domain from S. pombe | | Descriptor: | Putative Rho GTPase-activating protein C1565.02c, TETRAETHYLENE GLYCOL | | Authors: | Chichili, V.P.R, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel intertwined anti-parallel dimeric structure of scaffold BCH domain regulates RhoA and RhoGAP functions
Proc.Natl.Acad.Sci.USA, 2021
|
|
5OKD
 
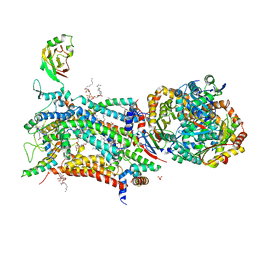 | | Crystal structure of bovine Cytochrome bc1 in complex with inhibitor SCR0911. | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, O'Neill, P.M, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2017-07-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
5OL5
 
 | | Crystal structure of an inactivated Ssp SICLOPPS intein with CFAHPQ extein | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, DNA polymerase III subunit alpha,DNA polymerase III subunit alpha, ... | | Authors: | Kick, L.M, Schneider, S. | | Deposit date: | 2017-07-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Mechanistic Insights into Cyclic Peptide Generation by DnaE Split-Inteins through Quantitative and Structural Investigation.
Chembiochem, 18, 2017
|
|
2PEB
 
 | |
7XYO
 
 | | Crystal Structure of a M61 aminopeptidase family member from Myxococcus fulvus | | Descriptor: | Aminopeptidase M61, HEXAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chen, X, Wang, X, Huo, L, Wu, D. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Characterization of a Myxobacterial Lanthipeptide with Unique Biosynthetic Features and Anti-inflammatory Activity.
J.Am.Chem.Soc., 145, 2023
|
|
