6E2V
 
 | | MDDEF in complex with MVAPP, ADPBeF3 and magnesium | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Stauffacher, C.V, Chen, C.-L. | | Deposit date: | 2018-07-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Visualizing the enzyme mechanism of mevalonate diphosphate decarboxylase.
Nat Commun, 11, 2020
|
|
6E2Y
 
 | | MDDEF in complex with MVAPP, ADP, sulfate and cobalt. Anomalous data | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Stauffacher, C.V, Chen, C.-L. | | Deposit date: | 2018-07-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Visualizing the enzyme mechanism of mevalonate diphosphate decarboxylase.
Nat Commun, 11, 2020
|
|
6E2Q
 
 | |
1R4I
 
 | | Crystal Structure of Androgen Receptor DNA-Binding Domain Bound to a Direct Repeat Response Element | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*CP*AP*AP*GP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*CP*TP*TP*GP*AP*TP*GP*TP*TP*CP*TP*GP*G)-3', Androgen receptor, ... | | Authors: | Shaffer, P.L, Jivan, A, Dollins, D.E, Claessens, F, Gewirth, D.T. | | Deposit date: | 2003-10-06 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of androgen receptor binding to selective androgen response elements.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1BO5
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN ESCHERICHIA COLI GLYCEROL KINASE AND THE ALLOSTERIC REGULATOR FRUCTOSE 1,6-BISPHOSPHATE. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, PROTEIN (GLYCEROL KINASE) | | Authors: | Ormo, M, Bystrom, C.E, Remington, S.J. | | Deposit date: | 1998-08-10 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate.
Biochemistry, 37, 1998
|
|
1BOT
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN ESCHERICHIA COLI GLYCEROL KINASE AND THE ALLOSTERIC REGULATOR FRUCTOSE 1,6-BISPHOSPHATE. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, PROTEIN (GLYCEROL KINASE) | | Authors: | Ormo, M, Bystrom, C.E, Remington, S.J. | | Deposit date: | 1998-08-05 | | Release date: | 1999-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate.
Biochemistry, 37, 1998
|
|
2RDS
 
 | | Crystal Structure of PtlH with Fe/oxalylglycine and ent-1-deoxypentalenic acid bound | | Descriptor: | (1S,3aS,5aR,8aS)-1,7,7-trimethyl-1,2,3,3a,5a,6,7,8-octahydrocyclopenta[c]pentalene-4-carboxylic acid, 1-deoxypentalenic acid 11-beta hydroxylase; Fe(II)/alpha-ketoglutarate dependent hydroxylase, FE (III) ION, ... | | Authors: | You, Z, Omura, S, Ikeda, H, Cane, D.E, Jogl, G. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Non-heme Iron Dioxygenase PtlH in Pentalenolactone Biosynthesis.
J.Biol.Chem., 282, 2007
|
|
6EJW
 
 | | Tryptophan Repressor TrpR from E.coli wildtype with Indole-3-acetic acid as ligand | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Trp operon repressor | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-23 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
4UT6
 
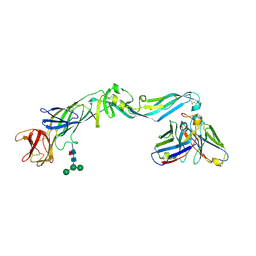 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 B7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 B7, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4V14
 
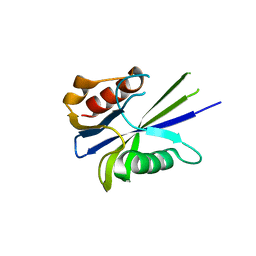 | |
6EKT
 
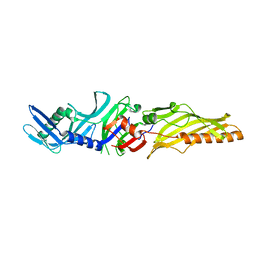 | |
1WM4
 
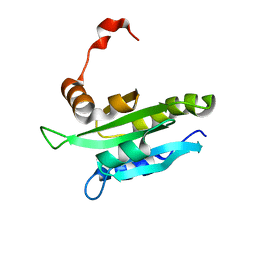 | | Solution structure of mouse coactosin, an actin filament binding protein | | Descriptor: | Coactosin-like protein | | Authors: | Hellman, M, Paavilainen, V.O, Naumanen, P, Lappalainen, P, Annila, A, Permi, P. | | Deposit date: | 2004-07-03 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of coactosin reveals structural homology to ADF/cofilin family proteins
Febs Lett., 576, 2004
|
|
4V2A
 
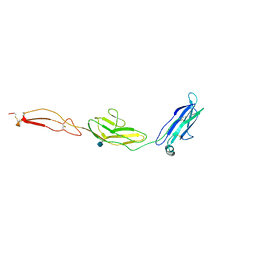 | | human Unc5A ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NETRIN RECEPTOR UNC5A | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
2RI1
 
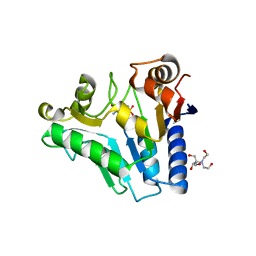 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) with GlcN6P from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine-6-phosphate deaminase | | Authors: | Liu, C, Li, D, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
4V42
 
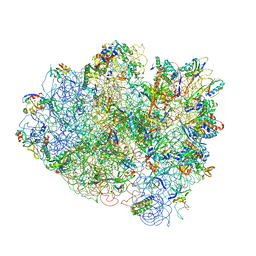 | | Crystal structure of the ribosome at 5.5 A resolution. | | Descriptor: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yusupov, M.M, Yusupova, G.Z, Baucom, A, Lieberman, K, Earnest, T.N, Cate, J.H.D, Noller, H.F. | | Deposit date: | 2001-03-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal structure of the ribosome at 5.5 A resolution
Science, 292, 2001
|
|
2M3O
 
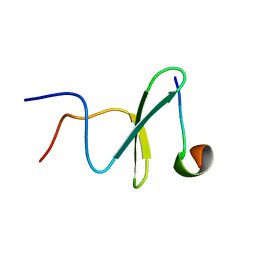 | | Structure and dynamics of a human Nedd4 WW domain-ENaC complex | | Descriptor: | Amiloride-sensitive sodium channel subunit alpha, E3 ubiquitin-protein ligase NEDD4 | | Authors: | Bobby, R, Medini, K, Neudecker, P, Lee, V, MacDonald, F.J, Brimble, M.A, Lott, J, Dingley, A.J. | | Deposit date: | 2013-01-23 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of human Nedd4-1 WW3 in complex with the alpha ENaC PY motif.
Biochim.Biophys.Acta, 1834, 2013
|
|
2RSY
 
 | | Solution structure of the SH2 domain of Csk in complex with a phosphopeptide from Cbp | | Descriptor: | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1, Tyrosine-protein kinase CSK | | Authors: | Tanaka, H, Akagi, K, Oneyama, C, Tanaka, M, Sasaki, Y, Kanou, T, Lee, Y, Yokogawa, D, Debenecker, M, Nakagawa, A, Okada, M, Ikegami, T. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-10 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Identification of a new interaction mode between the Src homology 2 domain of C-terminal Src kinase (Csk) and Csk-binding protein/phosphoprotein associated with glycosphingolipid microdomains.
J.Biol.Chem., 288, 2013
|
|
6EQB
 
 | | HLA class I histocompatibility antigen | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALA-ALA-GLY-ILE-GLY-ILE-LEU-THR-VAL, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | TCR-induced alteration of primary MHC peptide anchor residue.
Eur.J.Immunol., 49, 2019
|
|
6E5G
 
 | |
4V99
 
 | |
6EQG
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P21 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2RUX
 
 | | Solution structures of the DNA-binding domain (ZF6) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RV4
 
 | | Solution structures of the DNA-binding domain (ZF5) of mouse immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
6E5Y
 
 | | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP.
To Be Published
|
|
8IZE
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 4-HMBPP | | Descriptor: | Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, [(E)-3-(hydroxymethyl)pent-2-enyl] phosphono hydrogen phosphate | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
