7AZS
 
 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-569 | | Descriptor: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,6R,E)-3-hydroxy-4-(methoxyimino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | Authors: | Jenner, L.B, Yusupov, M, Yusupova, G, Rak, A. | | Deposit date: | 2020-11-17 | | Release date: | 2022-06-08 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
7TU4
 
 | | Structure of the L. blandensis dGTPase del55-58 mutant bound to Mn | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Sikkema, A.P, Klemm, B.P, Horng, J.C, Hall, T.M.T. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7OGK
 
 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ) | | Descriptor: | 3'ETS(LeuZ), Polyribonucleotide nucleotidyltransferase | | Authors: | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
7AZO
 
 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-977 | | Descriptor: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,4R,6R)-3-hydroxy-4-(methoxyamino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-((2-nitrophenyl)sulfonamido)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | Authors: | Jenner, L.B, Yusupov, M, Yusupova, G. | | Deposit date: | 2020-11-17 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
8C34
 
 | |
7P2T
 
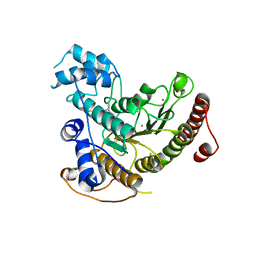 | | Tetartohedrally twinned crystal structure of Schistosoma mansoni HDAC8 in complex with a tricyclic thieno[3,2-b]indole capped hydroxamate-based inhibitor, bromine derivative | | Descriptor: | 5-[[(2R)-7-bromanyl-2-phenyl-2,3-dihydrothieno[3,2-b]indol-4-yl]methyl]-N-oxidanyl-thiophene-2-carboxamide, CHLORIDE ION, Histone deacetylase 8, ... | | Authors: | Saccoccia, F, Gemma, S, Campiani, G, Ruberti, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Schistosoma mansoni histone deacetylase 8 reveal a novel binding site for allosteric inhibitors.
J.Biol.Chem., 298, 2022
|
|
6HAX
 
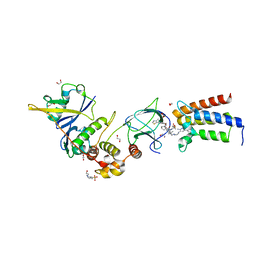 | | Crystal structure of PROTAC 2 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
6ZUL
 
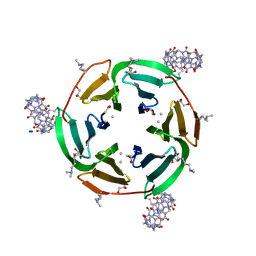 | | Crystal structure of dimethylated RSL in complex with cucurbit[7]uril and zinc | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SODIUM ION, ... | | Authors: | Guagnini, F, Engilberge, S, Flood, R.J, Ramberg, K.O, Crowley, P.B. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Metal-Mediated Protein-Cucurbituril Crystalline Architectures
Cryst.Growth Des., 2020
|
|
8QP0
 
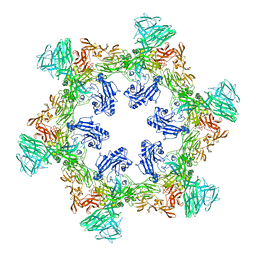 | | A hexamer pore in the S-layer of Sulfolobus acidocaldarius formed by SlaA protein | | Descriptor: | S-layer protein A | | Authors: | Gambelli, L, McLaren, M, Isupov, M, Conners, R, Daum, B. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.2 Å) | | Cite: | Structure of the two-component S-layer of the archaeon Sulfolobus acidocaldarius.
Elife, 13, 2024
|
|
6VPE
 
 | |
5JZR
 
 | | Solid-state MAS NMR structure of Acinetobacter phage 205 (AP205) coat protein in assembled capsid particles | | Descriptor: | Coat protein | | Authors: | Jaudzems, K, Andreas, L.B, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6YVP
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) complexed with dGMP and refined to 2.77 A | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, Histidine triad nucleotide-binding protein 2, mitochondrial | | Authors: | Dolot, R.D, Krakowiak, A, Nawrot, B.C. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6QL5
 
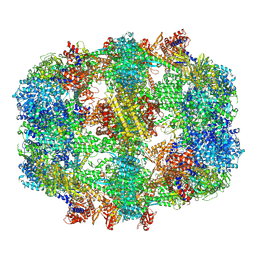 | | Structure of fatty acid synthase complex with bound gamma subunit from Saccharomyces cerevisiae at 2.8 angstrom | | Descriptor: | 4'-PHOSPHOPANTETHEINE, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
6ZUM
 
 | | Crystal structure of dimethylated RSL-N23H (RSL-B3) in complex with cucurbit[7]uril and zinc | | Descriptor: | ACETATE ION, Fucose-binding lectin protein, SODIUM ION, ... | | Authors: | Guagnini, F, Engilberge, S, Flood, R.J, Ramberg, K.O, Crowley, P.B. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Metal-Mediated Protein-Cucurbituril Crystalline Architectures
Cryst.Growth Des., 2020
|
|
5LQ1
 
 | | 1.41 A resolution structure of PtxB from Trichodesmium erythraeum IMS101 in complex with methylphosphonate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, METHYLPHOSPHONIC ACID ESTER GROUP, ... | | Authors: | Bisson, C, Adams, N.B.P, Polyviou, D, Bibby, T.S, Hunter, C.N, Hitchcock, A. | | Deposit date: | 2016-08-15 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The molecular basis of phosphite and hypophosphite recognition by ABC-transporters.
Nat Commun, 8, 2017
|
|
8RDZ
 
 | |
5K1I
 
 | | PDE4 crystal structure in complex with small molecule inhibitor | | Descriptor: | 4-[(5-acetyl-2-ethyl-3-oxo-6-phenyl-2,3-dihydropyridazin-4-yl)amino]benzoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Segarra, V, Hernandez, B, Ferrer-Miralles, N, Korndoerfer, I, Aymami, J. | | Deposit date: | 2016-05-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Biphenyl Pyridazinone Derivatives as Inhaled PDE4 Inhibitors: Structural Biology and Structure-Activity Relationships.
J. Med. Chem., 59, 2016
|
|
7RDX
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - open class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5K6G
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-24 DS-Cav1 variant. | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0 | | Authors: | Joyce, M.G, Zhang, B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5LRG
 
 | | Crystal structure of the porcine carboxypeptidase B - Anabaenopeptin B complex | | Descriptor: | Anabaenopeptin B, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Isolation, Co-Crystallization and Structure-Based Characterization of Anabaenopeptins as Highly Potent Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Sci Rep, 6, 2016
|
|
6VM8
 
 | |
5K9D
 
 | | Crystal structure of human dihydroorotate dehydrogenase at 1.7 A resolution | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lewis, T.A, Sykes, D.B, Law, J.M, Munoz, B, Scadden, D.T, Rustiguel, J.K, Nonato, M.C, Schreiber, S.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia.
ACS Med Chem Lett, 7, 2016
|
|
6HJP
 
 | |
7V35
 
 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.h, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
6VS6
 
 | |
