8F1C
 
 | | Voltage-gated potassium channel Kv3.1 with novel positive modulator (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one (compound 4) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (9M)-9-{5-chloro-6-[(3,3-dimethyl-2,3-dihydro-1-benzofuran-4-yl)oxy]-4-methylpyridin-3-yl}-2-methyl-7,9-dihydro-8H-purin-8-one, POTASSIUM ION, ... | | Authors: | Chen, Y, Ishchenko, A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Identification and structural and biophysical characterization of a positive modulator of human Kv3.1 channels.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9QH0
 
 | |
8GHF
 
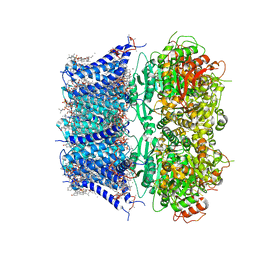 | | cryo-EM structure of hSlo1 in plasma membrane vesicles | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Tao, X, Zhao, C, MacKinnon, R. | | Deposit date: | 2023-03-10 | | Release date: | 2023-05-10 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Membrane protein isolation and structure determination in cell-derived membrane vesicles.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3OLZ
 
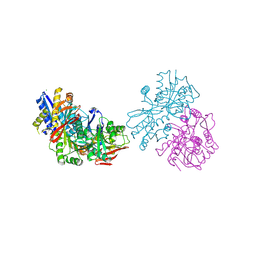 | |
3OM0
 
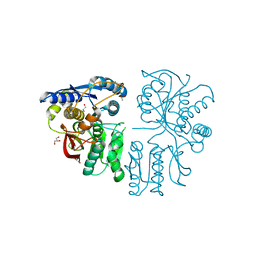 | |
2XUT
 
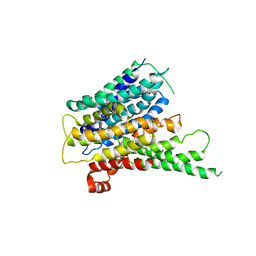 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
3QLV
 
 | |
3QLT
 
 | |
3QLU
 
 | |
7OY5
 
 | | Crystal structure of GSK3Beta in complex with ARN25068 | | Descriptor: | CHLORIDE ION, Glycogen synthase kinase-3 beta, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | Authors: | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Giabbai, B, Storici, P, Ortega, J, Girotto, S, Cavalli, A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
7OY6
 
 | | Crystal structure of human DYRK1A in complex with ARN25068 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | Authors: | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Ortega, J, Girotto, S, Cavalli, A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
6EI3
 
 | | Crystal structure of auto inhibited POT family peptide transporter | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Proton-dependent oligopeptide transporter family protein | | Authors: | Newstead, S, Brinth, A, Vogeley, L, Caffrey, M. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proton movement and coupling in the POT family of peptide transporters.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
9BH5
 
 | | High-resolution C. elegans 80S ribosome structure - class 1 | | Descriptor: | 18S rRNA, 28S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Sehgal, E, Serrao, V.H.B, Arribere, J. | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | High-resolution reconstruction of a C. elegans ribosome sheds light on evolutionary dynamics and tissue specificity.
Rna, 30, 2024
|
|
6LQM
 
 | | Cryo-EM structure of a pre-60S ribosomal subunit - state C | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-14 | | Release date: | 2020-08-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
4V02
 
 | | MinC:MinD cell division protein complex, Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROBABLE SEPTUM SITE-DETERMINING PROTEIN MINC, ... | | Authors: | Ghosal, D, Lowe, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mincd Cell Division Proteins Form Alternating Copolymeric Cytomotive Filaments.
Nat.Commun., 5, 2014
|
|
6A62
 
 | | Placental protein 13/galectin-13 variant R53HH57RD33G with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A64
 
 | | Placental protein 13/galectin-13 variant R53HR55NH57RD33G with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A63
 
 | | Placental protein 13/galectin-13 variant R53HH57R with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A66
 
 | | Placental protein 13/galectin-13 variant R53H with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J.Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
9C6V
 
 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (1 molecule/asymmetric unit) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
9C6X
 
 | | Crystal Structure of a single chain trimer composed of HLA-B*39:01 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, NACHT, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
9C6W
 
 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (2 molecules/asymmetric unit) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
9CAI
 
 | | High-resolution C. elegans 80S ribosome structure - class 1 | | Descriptor: | 18S rRNA, 28S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Sehgal, E, Serrao, V.H.B, Arribere, J. | | Deposit date: | 2024-06-17 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | High-resolution reconstruction of a C. elegans ribosome sheds light on evolutionary dynamics and tissue specificity.
Rna, 30, 2024
|
|
6LSR
 
 | | Cryo-EM structure of a pre-60S ribosomal subunit - state B | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-20 | | Release date: | 2020-08-26 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6A65
 
 | | Placental protein 13/galectin-13 variant R53HR55N with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
