6ZGZ
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6OAC
 
 | | PQR530 [(S)-4-(Difluoromethyl)-5-(4-(3-methylmorpholino)-6-morpholino-1,3,5-triazin-2-yl)pyridin-2-amine] bound to the PI3Ka catalytic subunit p110alpha | | Descriptor: | 4-(difluoromethyl)-5-{4-[(3S)-3-methylmorpholin-4-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, McPhail, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | (S)-4-(Difluoromethyl)-5-(4-(3-methylmorpholino)-6-morpholino-1,3,5-triazin-2-yl)pyridin-2-amine (PQR530), a Potent, Orally Bioavailable, and Brain-Penetrable Dual Inhibitor of Class I PI3K and mTOR Kinase.
J.Med.Chem., 62, 2019
|
|
8G5X
 
 | | Structure of the Class II Fructose-1,6-Bisphophatase from Francisella tularensis complexed with native metal cofactor Mn++ and substrate Fructose-1,6-Bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, GLYCEROL, ... | | Authors: | Abad-Zapatero, C, Selezneva, A.I. | | Deposit date: | 2023-02-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | New structures of Class II Fructose-1,6-Bisphosphatase from Francisella tularensis provide a framework for a novel catalytic mechanism for the entire class.
Plos One, 18, 2023
|
|
8FUJ
 
 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
6PJB
 
 | | HIV-1 Protease NL4-3 WT in Complex with Lopinavir | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
8HE7
 
 | | ADP-ribosyltransferase 1 (PARP1) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1, processed C-terminus, ... | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
6PJO
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-42 | | Descriptor: | Protease NL4-3, SULFATE ION, methyl [(1S)-1-cyclopentyl-2-({(2S,4S,5S)-5-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-4-hydroxy-1,6-diphenylhexan-2-yl}amino)-2-oxoethyl]carbamate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6NDR
 
 | |
7LK1
 
 | | Ornithine Aminotransferase (OAT) with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) - 1 Hour Soaking | | Descriptor: | (1R,4R)-4-fluoro-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-2-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Shen, S, Liu, D, Silverman, R. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
6WBG
 
 | | Cryo-EM structure of human Pannexin 1 channel with its C-terminal tail cleaved by caspase-7 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
7MLR
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(3,5-dimethylphenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1- yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (DW34) | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(3,5-dimethylphenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
7MLQ
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (compound 26) | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Johnson, J.A, Vail, N.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
7M3S
 
 | | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the alpha-and beta-site, sodium ion at the metal coordination site, and another F6F molecule at the enzyme beta-site at 1.55 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, CHLORIDE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-03-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the alpha-and beta-site, sodium ion at the metal coordination site, and another F6F molecule at the enzyme beta-site at 1.55 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
To be Published
|
|
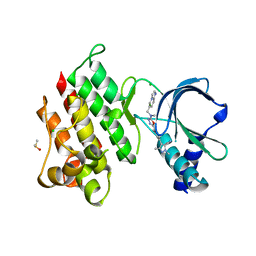
7ME8
 
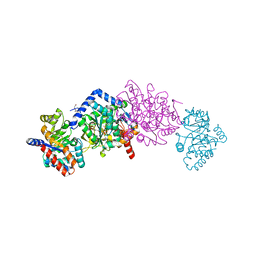 | | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the beta-site, sodium ion at the metal coordination site and dual beta-Q114 rotamer conformation at 1.60 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the beta-site, sodium ion at the metal coordination site and dual beta-Q114 rotamer conformation at 1.60 Angstrom resolution.
To be Published
|
|
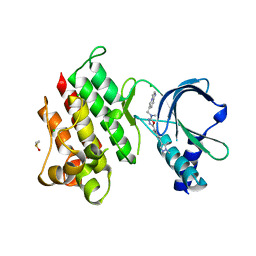
7OLV
 
 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl, cyano pyrazole group | | Descriptor: | 4-[4-[5-[imidazo[1,2-a]pyridin-6-ylmethyl(methyl)amino]-1,3,4-oxadiazol-2-yl]-3-methyl-phenyl]-2,5-dimethyl-pyrazole-3-carbonitrile, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
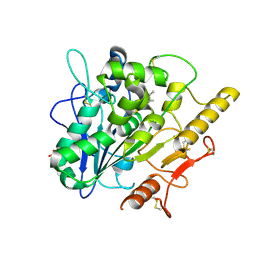
7OLS
 
 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl pyrazole group | | Descriptor: | 5-[4-(1,5-dimethylpyrazol-4-yl)-2-methyl-phenyl]-~{N}-(imidazo[1,2-a]pyridin-6-ylmethyl)-~{N}-methyl-1,3,4-oxadiazol-2-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
6YUW
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 454 | | Descriptor: | 1-(cyclopropylmethyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6ZGW
 
 | | Structure of human galactokinase 1 bound with (4-chlorophenyl)methyl pyridine-3-carboxylate | | Descriptor: | (4-chlorophenyl)methyl pyridine-3-carboxylate, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6OOT
 
 | | HIV-1 Protease NL4-3 L89V, L90M Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
7OSO
 
 | | The crystal structure of Erwinia tasmaniensis levansucrase in complex with (S)-1,2,4-butanentriol | | Descriptor: | (2~{S})-butane-1,2,4-triol, Levansucrase (Beta-D-fructofuranosyl transferase), ZINC ION | | Authors: | Polsinelli, I, Salomone-Stagni, M, Benini, S. | | Deposit date: | 2021-06-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Erwinia tasmaniensis levansucrase shows enantiomer selection for (S)-1,2,4-butanetriol.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6OPU
 
 | | HIV-1 Protease NL4-3 K45I, M46I, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPY
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, A71V, L76V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPS
 
 | | HIV-1 Protease NL4-3 WT in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPW
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, A71V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6ZH0
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
