4KSG
 
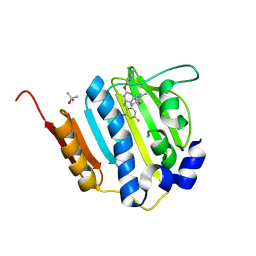 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor (4-[(1S,5R,6R)-6-AMINO-1-METHYL-3-AZABICYCLO[3.2.0]HEPT-3-YL]-6-FLUORO-N-METHYL-2-[(2-METHYLPYRIMIDIN-5-YL)OXY]-9H-PYRIMIDO[4,5-B]INDOL-8-AMINE) | | Descriptor: | 4-[(1S,5R,6R)-6-amino-1-methyl-3-azabicyclo[3.2.0]hept-3-yl]-6-fluoro-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | Authors: | Bensen, D.C, Akers-Rodriguez, S, Tari, L.W. | | Deposit date: | 2013-05-17 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A new class of type iia topoisomerase inhibitors with
broad-spectrum antibacterial activity
To be Published
|
|
4KTN
 
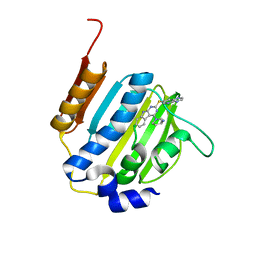 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor ((3S)-1-[2-(PYRIDO[2,3-B]PYRAZIN-7-YLSULFANYL)-9H-PYRIMIDO[4,5-B]INDOL-4-YL]PYRROLIDIN-3-AMINE) | | Descriptor: | (3S)-1-[2-(pyrido[2,3-b]pyrazin-7-ylsulfanyl)-9H-pyrimido[4,5-b]indol-4-yl]pyrrolidin-3-amine, DNA gyrase subunit B | | Authors: | Bensen, D.C, Akers-rodriguez, S, Tari, L.W. | | Deposit date: | 2013-05-20 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A new class of type iia topoisomerase inhibitors with
broad-spectrum antibacterial activity
To be Published
|
|
6CCW
 
 | |
1C11
 
 | | INTERCALATED D(TCCCGTTTCCA) DIMER, NMR, 7 STRUCTURES | | Descriptor: | DNA (5'-D(*TP*CP*CP*CP*GP*TP*TP*TP*CP*CP*A)-3') | | Authors: | Gallego, J, Chou, S.H, Reid, B.R. | | Deposit date: | 1998-07-15 | | Release date: | 1998-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Centromeric pyrimidine strands fold into an intercalated motif by forming a double hairpin with a novel T:G:G:T tetrad: solution structure of the d(TCCCGTTTCCA) dimer.
J.Mol.Biol., 273, 1997
|
|
6TQI
 
 | | I-MOTIF STRUCTURE FORMED FROM THE C STRAND OF A HUMAN TELOMERE FRAGMENT | | Descriptor: | DNA (5'-*TP*AP*AP*CP*CP*CP*TP*AP*A-3') | | Authors: | Parkinson, G.N, Wagner, A, Viladoms-Claverol, J, Duman, R, El-Omari, K. | | Deposit date: | 2019-12-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
2AZO
 
 | | DNA MISMATCH REPAIR PROTEIN MUTH FROM E. COLI | | Descriptor: | MUTH | | Authors: | Yang, W. | | Deposit date: | 1997-11-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases.
EMBO J., 17, 1998
|
|
2N1G
 
 | |
2N60
 
 | | G-quadruplexes with (4n-1) guanines in the G-tetrad core: formation of a G-triad water complex and implication for small-molecule binding | | Descriptor: | DNA (5'-D(*TP*TP*GP*TP*GP*TP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*GP*T)-3') | | Authors: | Heddi, B, Martin-Pintado, N, Serimbetov, Z, Kari, T.M, Phan, A.T. | | Deposit date: | 2015-08-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | G-quadruplexes with (4n - 1) guanines in the G-tetrad core: formation of a G-triadwater complex and implication for small-molecule binding
Nucleic Acids Res., 44, 2016
|
|
1PVG
 
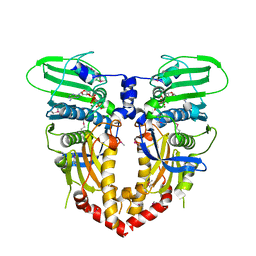 | |
2PIS
 
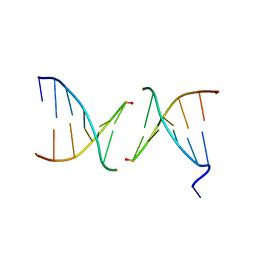 | | Efforts toward Expansion of the Genetic Alphabet: Structure and Replication of Unnatural Base Pairs | | Descriptor: | DNA (5'-D(*CP*GP*(CBR)P*GP*AP*AP*(FFD)P*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Matsuda, S, Fillo, J.D, Henry, A.A, Wilkins, S.J, Rai, P, Dwyer, T.J, Geierstanger, B.H, Wemmer, D.E, Schultz, P.G, Spraggon, G, Romesberg, F.E. | | Deposit date: | 2007-04-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Efforts toward expansion of the genetic alphabet: structure and replication of unnatural base pairs.
J.Am.Chem.Soc., 129, 2007
|
|
2F1I
 
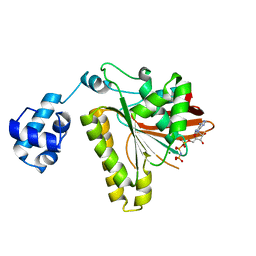 | | Recombinase in Complex with AMP-PNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Qian, X, He, Y, Wu, Y, Luo, Y. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae.
J.Mol.Biol., 360, 2006
|
|
2F1H
 
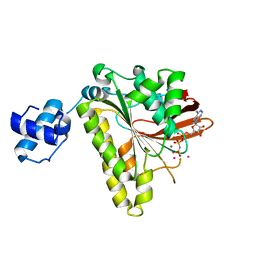 | | RECOMBINASE IN COMPLEX WITH AMP-PNP and Potassium | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Qian, X, He, Y, Wu, Y, Luo, Y. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae.
J.Mol.Biol., 360, 2006
|
|
2LFX
 
 | | Structure of the duplex when (5'S)-8,5'-cyclo-2'-deoxyguanosine is placed opposite dT | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*TP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of (5'S)-8,5'-Cyclo-2'-deoxyguanosine Mismatched with dA or dT.
Chem.Res.Toxicol., 25, 2012
|
|
2LFY
 
 | | Structure of the duplex when (5'S)-8,5'-cyclo-2'-deoxyguanosine is placed opposite dA | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of (5'S)-8,5'-Cyclo-2'-deoxyguanosine Mismatched with dA or dT.
Chem.Res.Toxicol., 25, 2012
|
|
6VSX
 
 | |
3IFF
 
 | | 2'-SeMe-A modified DNA decamer | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*GP*CP*GP*TP*(XUA)P*C)-3'), MAGNESIUM ION | | Authors: | Sheng, J, Gan, J, Salon, J, Huang, Z. | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selenium modification at 2'-position of adenosine for nucleic acid structure studies
To be Published
|
|
6WCK
 
 | | KRAS G-quadruplex G16T mutant with Bromo Uracil replacing T8 and T16. | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*CP*GP*GP*(BRU)P*GP*TP*GP*GP*GP*AP*AP*(BRU)P*AP*GP*GP*GP*AP*A)-3'), POTASSIUM ION | | Authors: | Schmidberger, J.W, Ou, A, Smith, N.M, Iyer, K.S, Bond, C.S. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | High resolution crystal structure of a KRAS promoter G-quadruplex reveals a dimer with extensive poly-A pi-stacking interactions for small-molecule recognition.
Nucleic Acids Res., 48, 2020
|
|
8I50
 
 | | Crystal structure of DNA octamer containing GuNA[Me,Me] | | Descriptor: | DNA (5'-D(*GP*(OIQ)P*GP*(BRU)P*AP*CP*AP*C)-3') | | Authors: | Aoyama, H, Obika, S, Yamaguchi, T. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Mechanism of the extremely high duplex-forming ability of oligonucleotides modified with N-tert-butylguanidine- or N-tert-butyl-N'- methylguanidine-bridged nucleic acids.
Nucleic Acids Res., 51, 2023
|
|
8HU5
 
 | | Crystal structure of DNA octamer containing GuNA[Me,tBu] | | Descriptor: | DNA (5'-D(*GP*(LR6)P*GP*(BRU)P*AP*CP*AP*C)-3') | | Authors: | Aoyama, H, Obika, H, Yamaguchi, T. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Mechanism of the extremely high duplex-forming ability of oligonucleotides modified with N-tert-butylguanidine- or N-tert-butyl-N'- methylguanidine-bridged nucleic acids.
Nucleic Acids Res., 51, 2023
|
|
8H1G
 
 | |
8H1F
 
 | |
8H1E
 
 | |
3AK8
 
 | | Crystal structure of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis | | Descriptor: | DNA protection during starvation protein, MAGNESIUM ION, SULFATE ION | | Authors: | Miyamoto, T, Asahina, Y, Miyazaki, S, Shimizu, H, Ohto, U, Noguchi, S, Satow, Y. | | Deposit date: | 2010-07-08 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2GDJ
 
 | | Delta-62 RADA recombinase in complex with AMP-PNP and magnesium | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, Y, Qian, X, He, Y, Luo, Y. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Rad51/RadA N-Terminal Domain Activates Nucleoprotein Filament ATPase Activity.
Structure, 14, 2006
|
|
3AK9
 
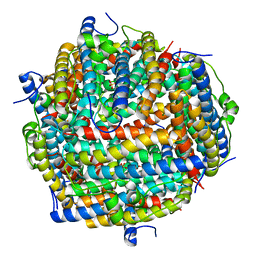 | | Crystal structure of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis, FE-soaked form | | Descriptor: | DNA protection during starvation protein, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Miyamoto, T, Asahina, Y, Miyazaki, S, Shimizu, H, Ohto, U, Noguchi, S, Satow, Y. | | Deposit date: | 2010-07-08 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis
Acta Crystallogr.,Sect.F, 67, 2011
|
|
