6P1S
 
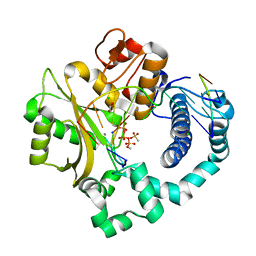 | | Post-catalytic nicked complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and newly incorporated AMP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
1HDD
 
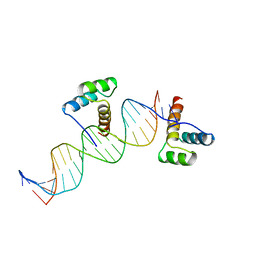 | | CRYSTAL STRUCTURE OF AN ENGRAILED HOMEODOMAIN-DNA COMPLEX AT 2.8 ANGSTROMS RESOLUTION: A FRAMEWORK FOR UNDERSTANDING HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*G P*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*C P*CP*TP*AP*A)-3'), PROTEIN (ENGRAILED HOMEODOMAIN) | | Authors: | Kissinger, C.R, Liu, B, Martin-Blanco, E, Kornberg, T.B, Pabo, C.O. | | Deposit date: | 1991-09-16 | | Release date: | 1992-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 63, 1990
|
|
5WNX
 
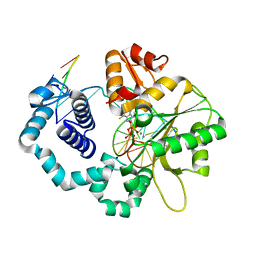 | | DNA polymerase beta substrate complex with incoming 6-TdGTP | | Descriptor: | 2-amino-9-{2-deoxy-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-1,9-dihydro-6H-purine-6-thione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
8EFK
 
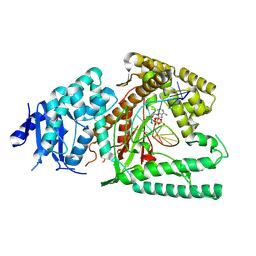 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with hairpin DNA | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(P*TP*TP*TP*TP*GP*GP*CP*TP*TP*TP*TP*GP*CP*CP*(2DA))-3'), Lates calcarifer DNA polymerase theta, ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
8EF9
 
 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3'), DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*C)-3'), ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
5WO0
 
 | | DNA polymerase beta substrate complex with incoming 5-FodUTP | | Descriptor: | 2'-deoxy-5-formyluridine 5'-(tetrahydrogen triphosphate), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
5WNZ
 
 | | DNA polymerase beta substrate complex with incoming 5-FodCTP | | Descriptor: | 2'-deoxy-5-formylcytidine 5'-(tetrahydrogen triphosphate), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
1HJC
 
 | |
5WNY
 
 | | DNA polymerase beta substrate complex with incoming 5-FdUTP | | Descriptor: | 2'-deoxy-5-fluorouridine 5'-(tetrahydrogen triphosphate), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schaich, M.A, Smith, M.R, Cloud, A.S, Holloran, S.M, Freudenthal, B.D. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of a DNA Polymerase Inserting Therapeutic Nucleotide Analogues.
Chem. Res. Toxicol., 30, 2017
|
|
6DIA
 
 | | DNA polymerase beta substrate complex with templating cytosine and incoming Fapy-dGTP analog | | Descriptor: | 1-[2-amino-5-(formylamino)-6-oxo-1,6-dihydropyrimidin-4-yl]-2,5-anhydro-1,3-dideoxy-6-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-ribo-hexitol, CALCIUM ION, DNA (5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Smith, M.R, Wilson, S.H, Beard, W.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | A guardian residue hinders insertion of a Fapy•dGTP analog by modulating the open-closed DNA polymerase transition.
Nucleic Acids Res., 47, 2019
|
|
8E24
 
 | | Human DNA polymerase theta in complex with allosteric inhibitor | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-[2,4-bis(trifluoromethyl)phenyl]-N-phenyl-N-[3-(pyridazin-3-yl)prop-2-yn-1-yl]acetamide, DNA, ... | | Authors: | Mader, P, Pau, V.P.T, Sicheri, F. | | Deposit date: | 2022-08-13 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Identification of RP-6685 , an Orally Bioavailable Compound that Inhibits the DNA Polymerase Activity of Pol theta.
J.Med.Chem., 65, 2022
|
|
8E23
 
 | | Human DNA polymerase theta in complex with allosteric inhibitor | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*GP*TP*CP*CP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*C)-3'), DNA (5'-D(*GP*C*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*G)-3'), ... | | Authors: | Mader, P, Pau, V.P.T, Sicheri, F. | | Deposit date: | 2022-08-13 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Identification of RP-6685 , an Orally Bioavailable Compound that Inhibits the DNA Polymerase Activity of Pol theta.
J.Med.Chem., 65, 2022
|
|
8V6G
 
 | | DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.16 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6H
 
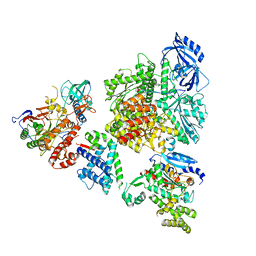 | | DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4JWM
 
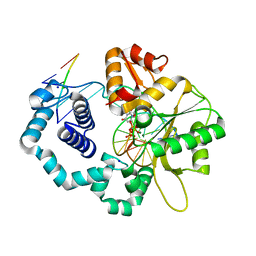 | | Ternary complex of D256E mutant of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Perera, L, Ping, L, Shock, D.D, Beard, W.A, Pedersen, L.C, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino Acid Substitution in the Active Site of DNA Polymerase beta Explains the Energy Barrier of the Nucleotidyl Transfer Reaction.
J.Am.Chem.Soc., 135, 2013
|
|
7OPL
 
 | |
8G9F
 
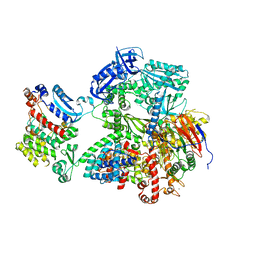 | | Complete auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3F27
 
 | | Structure of Sox17 Bound to DNA | | Descriptor: | DNA (5'-D(*DCP*DCP*DAP*DGP*DGP*DAP*DCP*DAP*DAP*DTP*DAP*DGP*DAP*DGP*DAP*DC)-3'), DNA (5'-D(*DGP*DTP*DCP*DTP*DCP*DTP*DAP*DTP*DTP*DGP*DTP*DCP*DCP*DTP*DGP*DG)-3'), Transcription factor SOX-17 | | Authors: | Palasingam, P, Jauch, R, Ng, C.K.L, Kolatkar, P.R. | | Deposit date: | 2008-10-29 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of Sox17 Bound to DNA Reveals a Conserved Bending Topology but Selective Protein Interaction Platforms
J.Mol.Biol., 388, 2009
|
|
3K3R
 
 | |
7K5P
 
 | | Bst DNA polymerase I time-resolved structure, 4 min post dATP addition | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Meza, R.A, Trinh, A.M, Chaput, J.C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Following replicative DNA synthesis by time-resolved X-ray crystallography.
Nat Commun, 12, 2021
|
|
6FBC
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification at the 3'-terminus of the primer. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBF
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the fourth primer nucleotide. | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*CP*GP*GP*TP*GP*CP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*GP*CP*AP*(OH3)P*CP*GP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8HT7
 
 | | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*TP*GP*GP*G)-3'), GLN-ALA-GLN-ALA-THR-ILE-SER-PHE-PRO-LYS-ARG-LYS-LEU-SER-TRP | | Authors: | Liu, C, Zhu, G, Geng, Y, Xu, N. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminal region of Cdc6 specifically recognizes human DNA G-quadruplex.
Int.J.Biol.Macromol., 260, 2024
|
|
6FBD
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the second primer nucleotide. | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*AP*(OH3)P*C)-3'), DNA (5'-D(P*AP*AP*AP*CP*GP*GP*TP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBG
 
 | | KlenTaq DNA polymerase processing a modified primer - bearing the modification upstream at the fifth primer nucleotide. | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*AP*AP*CP*GP*CP*CP*GP*GP*TP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Snapshots of a modified nucleotide moving through the confines of a DNA polymerase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
