7H0W
 
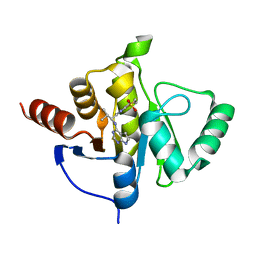 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013738-001 | | Descriptor: | 7-{(1R)-2-methyl-1-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]propyl}-3,4-dihydro-1lambda~6~-thiopyrano[2,3-b]pyridine-1,1(2H)-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.158 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6P3D
 
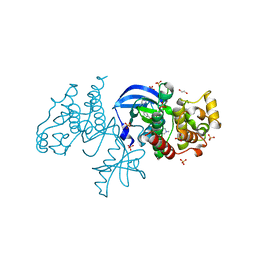 | | The co-crystal structure of BRAF(V600E) with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, AMMONIUM ION, ... | | Authors: | Agianian, B, Gavathiotis, E. | | Deposit date: | 2019-05-23 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Inhibitors of BRAF dimers using an allosteric site.
Nat Commun, 11, 2020
|
|
7NMS
 
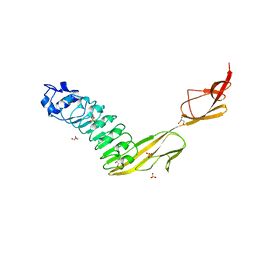 | |
8XQ1
 
 | | Crystal structure of spleen tyrosine kinase(SYK) in complex with SKI-G-1673 | | Descriptor: | Tyrosine-protein kinase SYK, cyclopropyl-[5-[[4-[4-[[(3~{R},4~{S})-3-methoxy-4-oxidanyl-pyrrolidin-1-yl]methyl]-3-methyl-pyrazol-1-yl]pyrimidin-2-yl]amino]-1-methyl-pyrrolo[2,3-b]pyridin-3-yl]methanone | | Authors: | Bong, S.M, Lee, B.I. | | Deposit date: | 2024-01-04 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of spleen tyrosine kinase(SYK) in complex with SKI-G-1673
To Be Published
|
|
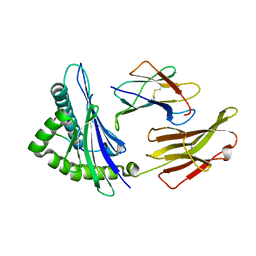
6MT3
 
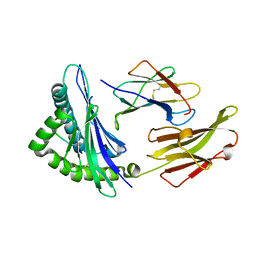 | |
6MT5
 
 | |
7HUV
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0031953-001 (A71EV2A-x3218) | | Descriptor: | (3S)-1-[(4S)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridine-4-carbonyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HUZ
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0032050-001 (A71EV2A-x3264) | | Descriptor: | 1-methyl-N-[2-(1,3-oxazol-4-yl)ethyl]-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.442 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
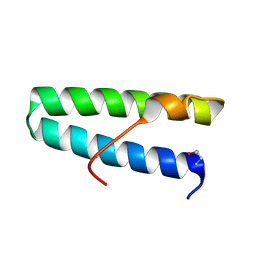
5VNY
 
 | |
7HWK
 
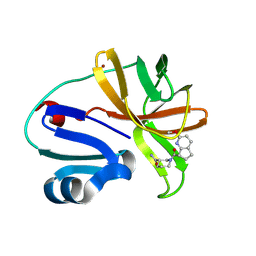 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0034455-001 (A71EV2A-x3954) | | Descriptor: | (3S)-1-[(7R)-2-methoxy-6,7-dihydro-5H-cyclopenta[b]pyridine-7-carbonyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7HXC
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0034391-001 (A71EV2A-x4239) | | Descriptor: | (3S,3aR,6aR)-1-[(2-chloro-6-methoxyphenyl)acetyl]octahydrocyclopenta[b]pyrrole-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-01-10 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6MT4
 
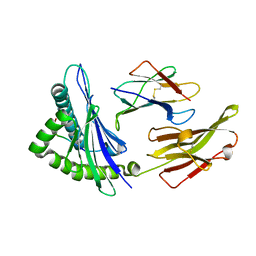 | |
7AJL
 
 | | Cyrstal structure of CYRI-B/Fam49B | | Descriptor: | CYFIP-related Rac1 interactor B | | Authors: | Yelland, T, Anh, H, Insall, R, Machesky, L, Ismail, S. | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Basis of CYRI-B Direct Competition with Scar/WAVE Complex for Rac1.
Structure, 29, 2021
|
|
8SCW
 
 | |
7I3Z
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0036444-001 (A71EV2A-x4606) | | Descriptor: | (3S)-1-[(7R)-5,7-dihydrothieno[3,4-b]pyridine-7-carbonyl]piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6OS7
 
 | |
7WPX
 
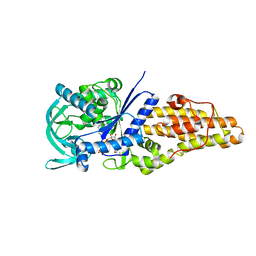 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a fragment and ATP | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranylimidazo[2,1-b][1,3]thiazole, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
6XKV
 
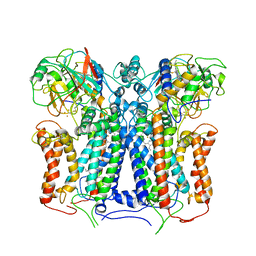 | | R. capsulatus cyt bc1 with both FeS proteins in b position (CIII2 b-b) | | Descriptor: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
7AVI
 
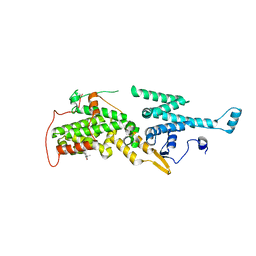 | | Crystal structure of SOS1 in complex with compound 2 | | Descriptor: | 3-propan-2-yl-~{N}-[(1~{R})-1-(3-sulfamoylphenyl)ethyl]-[1,2]oxazolo[5,4-b]pyridine-5-carboxamide, Son of sevenless homolog 1 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | One Atom Makes All the Difference: Getting a Foot in the Door between SOS1 and KRAS.
J.Med.Chem., 64, 2021
|
|
6V4T
 
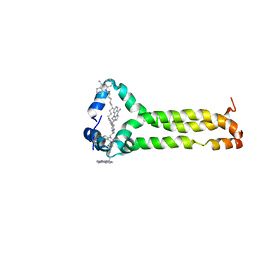 | | MPER-TMD of HIV-1 Env bound with the entry inhibitor S2C3 | | Descriptor: | 4,4'-(decane-1,10-diyl)bis(9-amino-2,3-dihydro-1H-cyclopenta[b]quinolin-4-ium), Envelope glycoprotein gp160 | | Authors: | Xiao, T, Frey, G, Fu, Q, Lavine, C.L, Scott, D.A, Seaman, M.S, Chou, J.J, Chen, B. | | Deposit date: | 2019-11-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 fusion inhibitors targeting the membrane-proximal external region of Env spikes.
Nat.Chem.Biol., 16, 2020
|
|
7USH
 
 | | BRD2-BD2 in complex with SF2523 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 2 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
2IXY
 
 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | Descriptor: | 5'-R(*GP*GP*CP*CP*UP*CP*CP*AP*AP*GP *CP*UP*GP*UP*GP*CP*CP*UP*UP*GP*GP*GP*UP*GP*GP*CP*C)-3' | | Authors: | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
6GWA
 
 | | Concanavalin B structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | Concanavalin B | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
9CHU
 
 | | Cryo-EM structure of calcineurin fused beta2 adrenergic receptor in norepinephrine bound inactive state | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, Beta-2 adrenergic receptor,Calcineurin subunit B type 1, Noradrenaline, ... | | Authors: | Xu, J, Chen, G, Du, Y, Kobilka, B.K. | | Deposit date: | 2024-07-02 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Calcineurin-fusion facilitates cryo-EM structure determination of a Family A GPCR.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9CHX
 
 | | cryo-EM structure of calcineurin-fused beta2 adrenergic receptor in carazolol bound inactive state | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, Beta-2 adrenergic receptor,Calcineurin subunit B type 1, ... | | Authors: | Xu, J, Chen, G, Du, Y, Kobilka, B.K. | | Deposit date: | 2024-07-02 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Calcineurin-fusion facilitates cryo-EM structure determination of a Family A GPCR.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
