5FYW
 
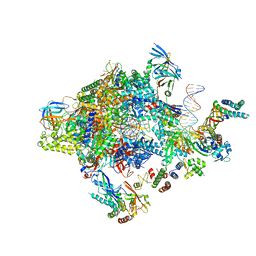 | | Transcription initiation complex structures elucidate DNA opening (OC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
8OJ8
 
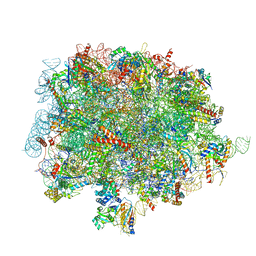 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ5
 
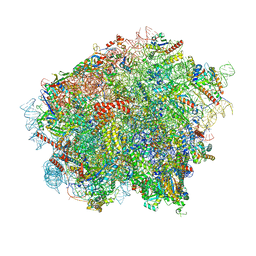 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Peter, J.J, Kulathu, Y, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
7TPT
 
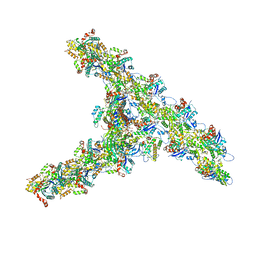 | | Single-particle Cryo-EM structure of Arp2/3 complex at branched-actin junction. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Ding, B, Narvaez-Ortiz, H.Y, Nolen, B.J, Chowdhury, S. | | Deposit date: | 2022-01-26 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of Arp2/3 complex at a branched actin filament junction resolved by single-particle cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8OKR
 
 | |
8AT2
 
 | | Structure of the augmin TIII subcomplex | | Descriptor: | HAUS augmin like complex subunit 4 L homeolog, HAUS augmin-like complex subunit 1, HAUS augmin-like complex subunit 3, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The augmin complex architecture reveals structural insights into microtubule branching.
Nat Commun, 13, 2022
|
|
8AT3
 
 | | Structure of the augmin holocomplex in open conformation | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, HAUS augmin like complex subunit 4 L homeolog, HAUS augmin like complex subunit 6 L homeolog, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (33 Å) | | Cite: | The augmin complex architecture reveals structural insights into microtubule branching.
Nat Commun, 13, 2022
|
|
8AT4
 
 | | Structure of the augmin holocomplex in closed conformation | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, HAUS augmin like complex subunit 4 L homeolog, HAUS augmin like complex subunit 6 L homeolog, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (33 Å) | | Cite: | The augmin complex architecture reveals structural insights into microtubule branching.
Nat Commun, 13, 2022
|
|
5CIQ
 
 | | Ran GDP wild type tetragonal crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
5CIW
 
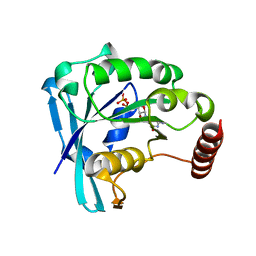 | | Ran GDP Y39A mutant monoclinic crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
5CIT
 
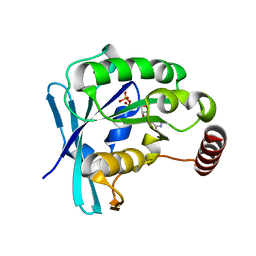 | | Ran GDP wild type monoclinic crystal form | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
7M12
 
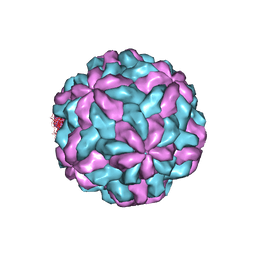 | | TVV2 capsid protein | | Descriptor: | Capsid protein | | Authors: | Zhou, H.Z, Stevens, A.W, Cui, Y.X, Muratore, K.A, Johnson, P.J. | | Deposit date: | 2021-03-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2.
Mbio, 12, 2021
|
|
7LWY
 
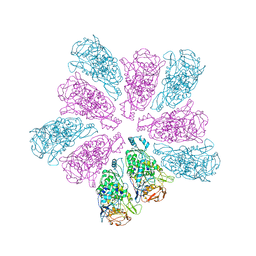 | | TVV viral capsid protein | | Descriptor: | Capsid protein | | Authors: | Zhou, Z.H, Stevens, A.W, Cui, Y.X, Johnson, P.J, Muratore, K.A. | | Deposit date: | 2021-03-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic Structure of the Trichomonas vaginalis Double-Stranded RNA Virus 2.
Mbio, 12, 2021
|
|
8CRO
 
 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Grunberger, F, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-08 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8R4I
 
 | |
7TXD
 
 | | Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Broadly neutralizing darpin bnd.9, ... | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DPG
 
 | | Cryo-EM structure of the 5HT2C receptor (INI isoform) bound to psilocin | | Descriptor: | 3-[2-(dimethylamino)ethyl]-1~{H}-indol-4-ol, 5-hydroxytryptamine receptor 2C, Antibody fragment scFv16, ... | | Authors: | Gumpper, R.H, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-07-15 | | Release date: | 2022-08-24 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular insights into the regulation of constitutive activity by RNA editing of 5HT 2C serotonin receptors.
Cell Rep, 40, 2022
|
|
7VIG
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DPI
 
 | | Cryo-EM structure of the 5HT2C receptor (VSV isoform) bound to lorcaserin | | Descriptor: | (1R)-8-chloro-1-methyl-2,3,4,5-tetrahydro-1H-3-benzazepine, 5-hydroxytryptamine receptor 2C, Antibody fragment scFv16, ... | | Authors: | Gumpper, R.H, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-07-15 | | Release date: | 2022-08-24 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into the regulation of constitutive activity by RNA editing of 5HT 2C serotonin receptors.
Cell Rep, 40, 2022
|
|
7ZL3
 
 | |
7VIE
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VIF
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with (S)-FTY720-P | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VIH
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DPF
 
 | | Cryo-EM structure of the 5HT2C receptor (INI isoform) bound to lorcaserin | | Descriptor: | (1R)-8-chloro-1-methyl-2,3,4,5-tetrahydro-1H-3-benzazepine, 5-hydroxytryptamine receptor 2C, Antibody fragment scFv16, ... | | Authors: | Gumpper, R.H, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-07-15 | | Release date: | 2022-08-24 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Molecular insights into the regulation of constitutive activity by RNA editing of 5HT 2C serotonin receptors.
Cell Rep, 40, 2022
|
|
8DPH
 
 | | Cryo-EM structure of the 5HT2C receptor (VGV isoform) bound to lorcaserin | | Descriptor: | (1R)-8-chloro-1-methyl-2,3,4,5-tetrahydro-1H-3-benzazepine, 5-hydroxytryptamine receptor 2C, Antibody fragment scFv16, ... | | Authors: | Gumpper, R.H, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-07-15 | | Release date: | 2022-08-24 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into the regulation of constitutive activity by RNA editing of 5HT 2C serotonin receptors.
Cell Rep, 40, 2022
|
|
