2LSU
 
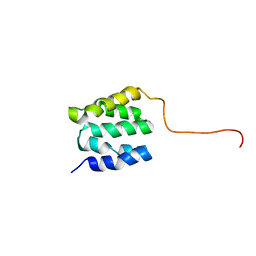 | | The NMR high resolution structure of yeast Tah1 in a free form | | Descriptor: | TPR repeat-containing protein associated with Hsp90 | | Authors: | Back, R, Dominguez, C, Rothe, B, Bobo, C, Beaufils, C, Morera, S, Meyer, P, Charpentier, B, Branlant, C, Allain, F, Manival, X. | | Deposit date: | 2012-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Analysis Shows How Tah1 Tethers Hsp90 to the R2TP Complex.
Structure, 21, 2013
|
|
2PWD
 
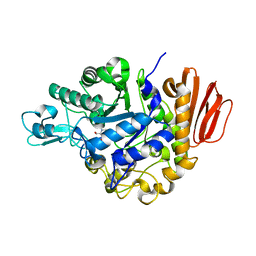 | | Crystal Structure of the Trehalulose Synthase MUTB from Pseudomonas Mesoacidophila MX-45 Complexed to the Inhibitor Deoxynojirmycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2PRG
 
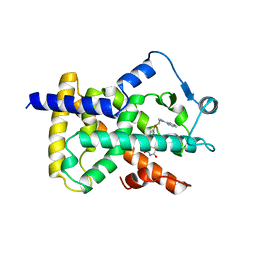 | | LIGAND-BINDING DOMAIN OF THE HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), NUCLEAR RECEPTOR COACTIVATOR SRC-1, ... | | Authors: | Nolte, R.T, Wisely, G.B, Milburn, M.V. | | Deposit date: | 1998-08-14 | | Release date: | 1999-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma.
Nature, 395, 1998
|
|
2PWF
 
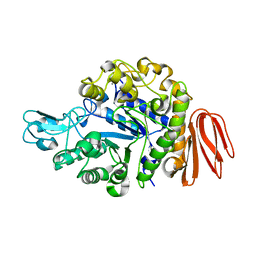 | | Crystal structure of the MutB D200A mutant in complex with glucose | | Descriptor: | CALCIUM ION, Sucrose isomerase, beta-D-glucopyranose | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2M50
 
 | | Analysis of the structural and molecular basis of voltage-sensitive sodium channel inhibition by the spider toxin, Huwentoxin-IV (-TRTX-Hh2a). | | Descriptor: | Mu-theraphotoxin-Hh2a | | Authors: | Gibbs, A, Minassian, N, Flinspach, M, Wickenden, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the Structural and Molecular Basis of Voltage-sensitive Sodium Channel Inhibition by the Spider Toxin Huwentoxin-IV ( mu-TRTX-Hh2a).
J.Biol.Chem., 288, 2013
|
|
2PWG
 
 | | Crystal Structure of the Trehalulose Synthase MutB From Pseudomonas Mesoacidophila MX-45 Complexed to the Inhibitor Castanospermine | | Descriptor: | CALCIUM ION, CASTANOSPERMINE, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
2PWE
 
 | | Crystal structure of the MutB E254Q mutant in complex with the substrate sucrose | | Descriptor: | CALCIUM ION, Sucrose isomerase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 282, 2007
|
|
3NR5
 
 | | Crystal structure of human Maf1 | | Descriptor: | Repressor of RNA polymerase III transcription MAF1 homolog | | Authors: | Ringel, R, Vannini, A, Kusser, A.G, Berninghausen, O, Kassavetis, G.A, Cramer, P. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular Basis of RNA Polymerase III Transcription Repression by Maf1
Cell(Cambridge,Mass.), 143, 2010
|
|
3P91
 
 | |
3ORY
 
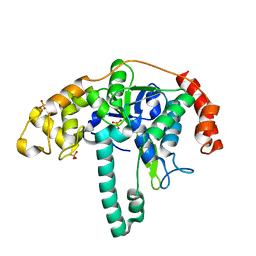 | | Crystal structure of Flap endonuclease 1 from hyperthermophilic archaeon Desulfurococcus amylolyticus | | Descriptor: | PHOSPHATE ION, flap endonuclease 1 | | Authors: | Mase, T, Kubota, K, Miyazono, K, Kawarabayashii, Y, Tanokura, M. | | Deposit date: | 2010-09-08 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of flap endonuclease 1 from the hyperthermophilic archaeon Desulfurococcus amylolyticus
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3P8B
 
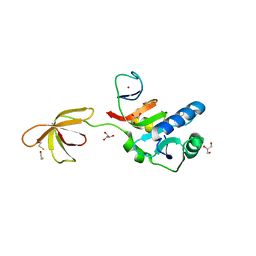 | |
3QFP
 
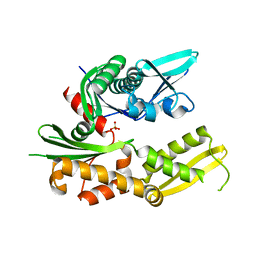 | | Crystal structure of yeast Hsp70 (Bip/Kar2) ATPase domain | | Descriptor: | 78 kDa glucose-regulated protein homolog, PHOSPHATE ION | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3QML
 
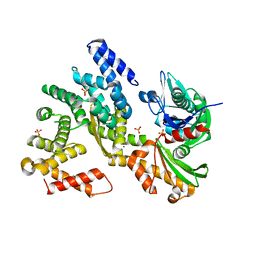 | | The structural analysis of Sil1-Bip complex reveals the mechanism for Sil1 to function as a novel nucleotide exchange factor | | Descriptor: | 78 kDa glucose-regulated protein homolog, MAGNESIUM ION, Nucleotide exchange factor SIL1, ... | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-02-04 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3QFU
 
 | | Crystal structure of Yeast Hsp70 (Bip/kar2) complexed with ADP | | Descriptor: | 78 kDa glucose-regulated protein homolog, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3P22
 
 | | Crystal structure of the ENE, a viral RNA stability element, in complex with A9 RNA | | Descriptor: | Core ENE hairpin from KSHV PAN RNA, oligo(A)9 RNA | | Authors: | Mitton-Fry, R.M, DeGregorio, S.J, Wang, J, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2010-10-01 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Poly(A) tail recognition by a viral RNA element through assembly of a triple helix.
Science, 330, 2010
|
|
3O44
 
 | |
3RRX
 
 | | Crystal Structure of Q683A mutant of Exo-1,3/1,4-beta-glucanase (ExoP) from Pseudoalteromonas sp. BB1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Exo-1,3/1,4-beta-glucanase, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2011-05-01 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain
Febs J., 279, 2012
|
|
3OLC
 
 | |
3RQR
 
 | | Crystal structure of the RYR domain of the rabbit ryanodine receptor | | Descriptor: | (UNK)(UNK)(UNK)(UNK), Ryanodine receptor 1 | | Authors: | Nair, U.B, Li, W, Dong, A, Walker, J.R, Gramolini, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-28 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural determination of the phosphorylation domain of the ryanodine receptor.
Febs J., 279, 2012
|
|
4HK1
 
 | |
5MEE
 
 | | Cyanothece lipoxygenase 2 (CspLOX2) variant - L304V | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Arachidonate 15-lipoxygenase, FE (III) ION, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
5MEG
 
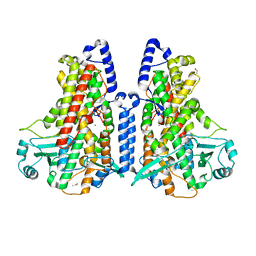 | | Manganese-substituted Cyanothece lipoxygenase 2 (Mn-CspLOX2) | | Descriptor: | Arachidonate 15-lipoxygenase, CHLORIDE ION, ETHANOL, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
5N61
 
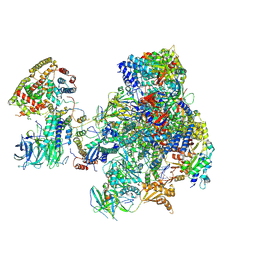 | | RNA polymerase I initially transcribing complex | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
5N7D
 
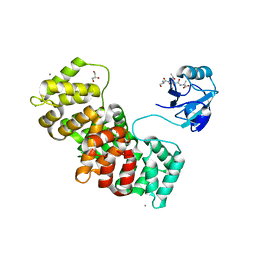 | | MAGI-1 complexed with a RSK1 peptide | | Descriptor: | CALCIUM ION, GLYCEROL, Membrane-associated guanylate kinase, ... | | Authors: | Gogl, G, Nyitray, L. | | Deposit date: | 2017-02-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamic control of RSK complexes by phosphoswitch-based regulation.
FEBS J., 285, 2018
|
|
5N5Y
 
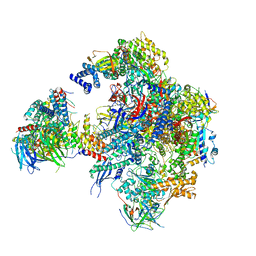 | | Cryo-EM structure of RNA polymerase I in complex with Rrn3 and Core Factor (Orientation III) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Engel, C, Gubbey, T, Neyer, S, Sainsbury, S, Oberthuer, C, Baejen, C, Bernecky, C, Cramer, P. | | Deposit date: | 2017-02-14 | | Release date: | 2017-04-05 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Basis of RNA Polymerase I Transcription Initiation.
Cell, 169, 2017
|
|
