6A3H
 
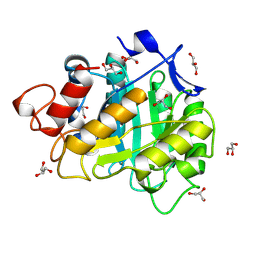 | | 2-DEOXY-2-FLURO-B-D-CELLOTRIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ENDOGLUCANASE, GLYCEROL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
4YFN
 
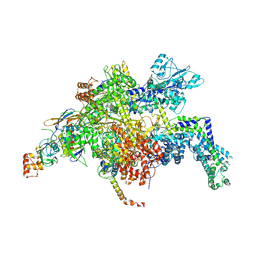 | | Escherichia coli RNA polymerase in complex with squaramide compound 14 (N-[3,4-dioxo-2-(4-{[4-(trifluoromethyl)benzyl]amino}piperidin-1-yl)cyclobut-1-en-1-yl]-3,5-dimethyl-1,2-oxazole-4-sulfonamide) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.817 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
7BVT
 
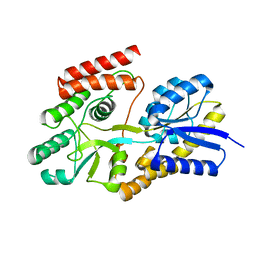 | | Crystal structure of cyclic alpha-maltosyl-1,6-maltose binding protein from Arthrobacter globiformis | | Descriptor: | Hypothetical sugar ABC-transporter sugar binding protein, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2020-04-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular analysis of cyclic alpha-maltosyl-(1→6)-maltose binding protein in the bacterial metabolic pathway.
Plos One, 15, 2020
|
|
7BXA
 
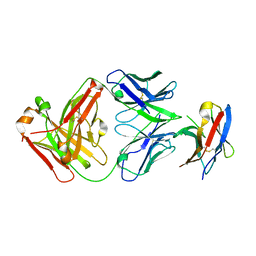 | | Crystal structure of PD-1 in complex with tislelizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S, Lee, S.H, Lim, H, Lee, H.T, Kim, Y.J, Park, E.B. | | Deposit date: | 2020-04-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal structure of PD-1 in complex with an antibody-drug tislelizumab used in tumor immune checkpoint therapy.
Biochem.Biophys.Res.Commun., 527, 2020
|
|
7BTS
 
 | | Structure of human beta1 adrenergic receptor bound to epinephrine and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-02 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
1NTV
 
 | | Crystal Structure of the Disabled-1 (Dab1) PTB domain-ApoER2 peptide complex | | Descriptor: | Apolipoprotein E Receptor-2 peptide, Disabled homolog 1, PHOSPHATE ION | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1DCY
 
 | |
4B5V
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with 4-hydroxyl-2-ketoheptane-1,7-dioate | | Descriptor: | (4R)-4-oxidanyl-2-oxidanylidene-heptanedioic acid, 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, GLYCEROL, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
1UAJ
 
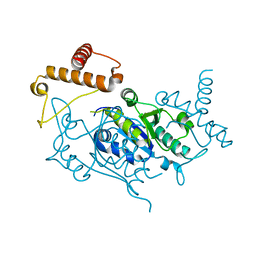 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
6TTA
 
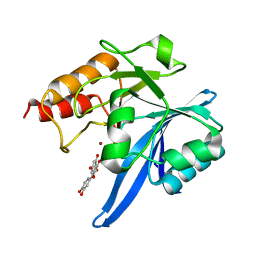 | | Haddock model of NDM-1/quercetin complex | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
3VQ4
 
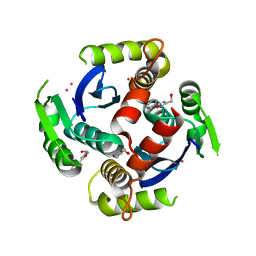 | | Fragments bound to HIV-1 integrase | | Descriptor: | (5-phenyl-1,2-oxazol-3-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
2Z4U
 
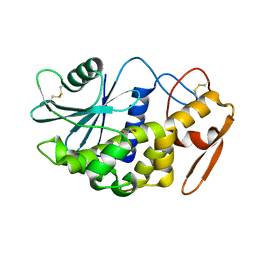 | |
6KE0
 
 | | Crystal structure of PDE10A in complex with a triazolopyrimidine inhibitor | | Descriptor: | 2-(5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)-1-[(2S)-2-methyl-1,2-dihydroimidazo[1,2-a]benzimidazol-3-yl]ethanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis, SAR study, and biological evaluation of novel 2,3-dihydro-1H-imidazo[1,2-a]benzimidazole derivatives as phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
4ZOB
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
6KK1
 
 | | Structure of thermal-stabilised(M8) human GLP-1 receptor transmembrane domain | | Descriptor: | Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
4ZQK
 
 | | Structure of the complex of human programmed death-1 (PD-1) and its ligand PD-L1. | | Descriptor: | Programmed cell death 1 ligand 1, Programmed cell death protein 1, SODIUM ION | | Authors: | Zak, K.M, Dubin, G, Holak, T.A. | | Deposit date: | 2015-05-10 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
5XOV
 
 | | Crystal structure of peptide-HLA-A24 bound to S19-2 V-delta/V-beta TCR | | Descriptor: | Beta-2-microglobulin, HIV-1 Nef138-10 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.684 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
6TOT
 
 | | Crystal structure of the Orexin-1 receptor in complex with lemborexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (1~{R},2~{S})-2-[(2,4-dimethylpyrimidin-5-yl)oxymethyl]-~{N}-(5-fluoranylpyridin-2-yl)-2-(3-fluorophenyl)cyclopropane-1-carboxamide, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
2NCS
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
6TOD
 
 | | Crystal structure of the Orexin-1 receptor in complex with EMPA | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
3VL8
 
 | | Crystal structure of XEG | | Descriptor: | SULFATE ION, Xyloglucan-specific endo-beta-1,4-glucanase A | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
6TQ9
 
 | | Crystal structure of the Orexin-1 receptor in complex with SB-408124 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-[6,8-bis(fluoranyl)-2-methyl-quinolin-4-yl]-3-[4-(dimethylamino)phenyl]urea, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
7XHG
 
 | |
5NVP
 
 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
4IWP
 
 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.065 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
