1HNC
 
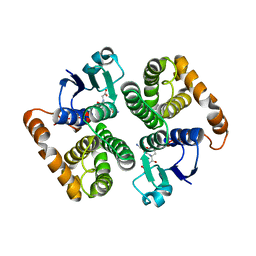 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
4PYY
 
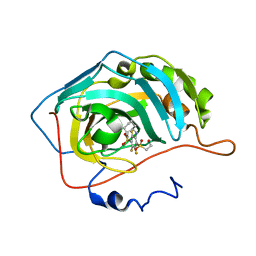 | | Crystal structure of human carbonic anhydrase isozyme II with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-03-28 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
3OYS
 
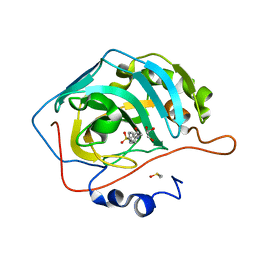 | | Human Carbonic Anhydrase II complexed with 2-{[4-AMINO-3-(3-HYDROXYPROP-1-YN-1-YL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-1-YL]METHYL}-5-METHYL-3-(2-METHYLPHENYL)QUINAZOLIN-4(3H)-ONE | | Descriptor: | 2-phenyl-N-(4-sulfamoylphenyl)acetamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Aggarwal, M, McKenna, R. | | Deposit date: | 2010-09-23 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Anticonvulsant 4-aminobenzenesulfonamide derivatives with branched-alkylamide moieties: X-ray crystallography and inhibition studies of human carbonic anhydrase isoforms I, II, VII, and XIV.
J.Med.Chem., 54, 2011
|
|
4JLG
 
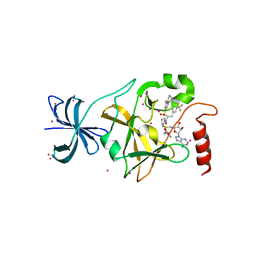 | | SETD7 in complex with inhibitor (R)-PFI-2 and S-adenosyl-methionine | | Descriptor: | 8-fluoro-N-{(2R)-1-oxo-1-(pyrrolidin-1-yl)-3-[3-(trifluoromethyl)phenyl]propan-2-yl}-1,2,3,4-tetrahydroisoquinoline-6-sulfonamide, Histone-lysine N-methyltransferase SETD7, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Wu, H, Zeng, H, El Bakkouri, M, Barsyte, D, Vedadi, M, Tatlock, J, Owen, D, Bunnage, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6R6Y
 
 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-chloro-2-cyclohexylsulfanyl-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-cyclohexylsulfanyl-~{N}-(2-hydroxyethyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
6V9N
 
 | |
1E40
 
 | | Tris/maltotriose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 2.2A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
1E43
 
 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.7A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
5J74
 
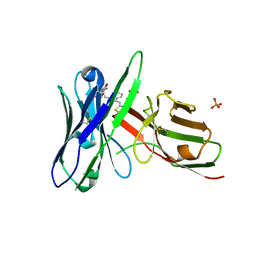 | | Fluorogen activating protein AM2.2 in complex with TO1-2p | | Descriptor: | 1-(17-amino-5,8-dioxo-12,15-dioxa-4,9-diazaheptadecan-1-yl)-4-{[3-(3-sulfopropyl)-1,3-benzothiazol-3-ium-2-yl]methyl}quinolin-1-ium, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
1E3X
 
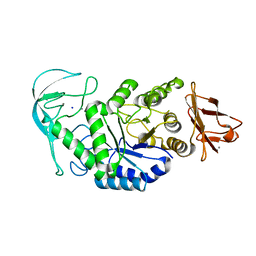 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.92A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-26 | | Release date: | 2001-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
5M26
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 in complex with methylhydroquinone | | Descriptor: | 2-methylbenzene-1,4-diol, FE (III) ION, Hydroquinone dioxygenase large subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5HFU
 
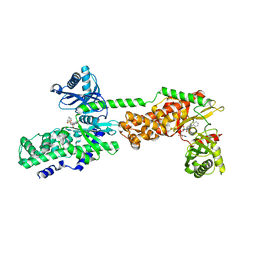 | | Crystal Structure of Human Hexokinase 2 with cmpd 27, a 2-amido-6-benzenesulfonamide glucosamine | | Descriptor: | Hexokinase-2, ~{N}-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-[[(4-cyanophenyl)sulfonylamino]methyl]-2,4,5-tris(oxidanyl)oxan-3-yl]-3-phenyl-benzamide | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
6SQU
 
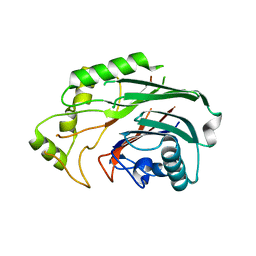 | | Crystal structure of human SHIP2 catalytic domain in complex with 1,2,4 Dimer | | Descriptor: | 5,5'-(ethane-1,2-diylbis(oxy))bis(benzene-5,4,2,1,-tetrayl)hexakisphosphate, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Whitfield, H, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2019-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Allosteric Site on SHIP2 Identified Through Fluorescent Ligand Screening and Crystallography: A Potential New Target for Intervention.
J.Med.Chem., 64, 2021
|
|
2EC6
 
 | | Placopecten Striated Muscle Myosin II | | Descriptor: | CALCIUM ION, Myosin essential light chain, Myosin heavy chain, ... | | Authors: | Yang, Y, Brown, J, Samudrala, G, Reutzel, R, Szent-Gyorgyi, A. | | Deposit date: | 2007-02-10 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
6R6F
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-chloro-2-cyclohexylsulfanyl-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 4-chloranyl-2-cyclohexylsulfanyl-~{N}-(2-hydroxyethyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-27 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
5TRK
 
 | |
7L3B
 
 | | T4 Lysozyme L99A - iodobenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3G
 
 | | T4 Lysozyme L99A - 4-iodotoluene - cryo | | Descriptor: | 1-iodo-4-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3F
 
 | | T4 Lysozyme L99A - 4-iodotoluene - RT | | Descriptor: | 1-iodo-4-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3E
 
 | | T4 Lysozyme L99A - 3-iodotoluene - cryo | | Descriptor: | 1-iodo-3-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3D
 
 | | T4 Lysozyme L99A - 3-iodotoluene - RT | | Descriptor: | 1-iodo-3-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3I
 
 | | T4 Lysozyme L99A - propylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endolysin, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
8WXB
 
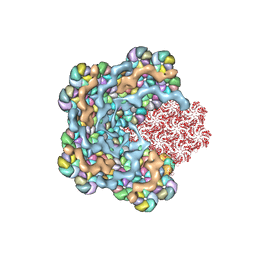 | | Cryo-EM structure of the alpha-carboxysome shell vertex from Prochlorococcus MED4 | | Descriptor: | Carboxysome assembly protein CsoS2, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1 | | Authors: | Jiang, Y.L, Zhou, R.Q, Zhou, C.Z, Zeng, Q.L. | | Deposit date: | 2023-10-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and assembly of the alpha-carboxysome in the marine cyanobacterium Prochlorococcus.
Nat.Plants, 10, 2024
|
|
2ANQ
 
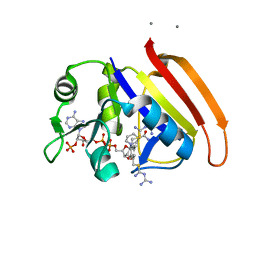 | | Crystal Structure of E.coli DHFR in complex with NADPH and the inhibitor compound 10a. | | Descriptor: | (2,5-dimethylbenzene-1,4-diyl)dimethanediyl bis(N-carbamimidoylcarbamimidothioate), Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
2AJC
 
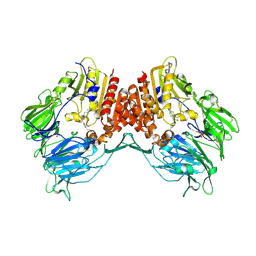 | | Porcine dipeptidyl peptidase IV (CD26) in complex with 4-(2-Aminoethyl)-benzene sulphonyl fluoride (AEBSF) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
