6USR
 
 | |
3KYL
 
 | |
6AR1
 
 | | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications (RT/Duplex (Nat)) | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA, GsI-IIC RT, ... | | Authors: | Stamos, J.L, Lentzsch, A.M, Lambowitz, A.M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications.
Mol. Cell, 68, 2017
|
|
6AR3
 
 | | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications (RT/Duplex (Se-Met)) | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA, GsI-IIC RT, ... | | Authors: | Stamos, J.L, Lentzsch, A.M, Lambowitz, A.M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications.
Mol. Cell, 68, 2017
|
|
8T2R
 
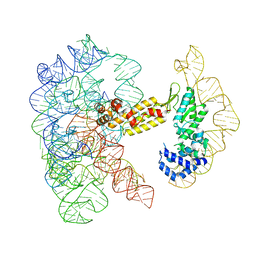 | | Structure of a group II intron ribonucleoprotein in the pre-ligation (pre-2F) state | | Descriptor: | 5'exon, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2T
 
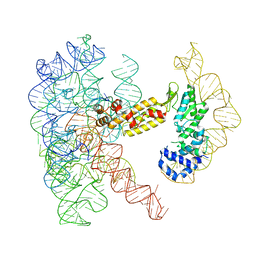 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | Descriptor: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2S
 
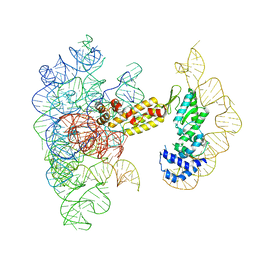 | | Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state | | Descriptor: | AMMONIUM ION, CALCIUM ION, Group II intron reverse transcriptase/maturase, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8SXU
 
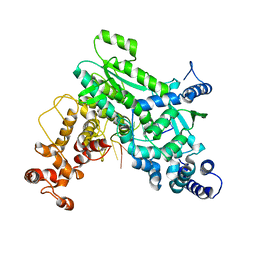 | |
8SXT
 
 | | Structure of LINE-1 ORF2p with template:primer hybrid | | Descriptor: | DNA primer, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | van Eeuwen, T, Taylor, M.S, Rout, M.P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
8UW3
 
 | | Human LINE-1 retrotransposon ORF2 protein engaged with template RNA in elongation state | | Descriptor: | Complementary DNA, LINE-1 retrotransposable element ORF2 protein, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Thawani, A, Florez Ariza, A.J, Collins, K, Nogales, E. | | Deposit date: | 2023-11-06 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Template and target-site recognition by human LINE-1 in retrotransposition.
Nature, 626, 2024
|
|
8WUT
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 2024
|
|
8WUS
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (40-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 2024
|
|
8WUV
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 2024
|
|
4O44
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in complex with 4-((4-(mesitylamino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ529), a non-nucleoside inhibitor | | Descriptor: | 4-({4-[3-(morpholin-4-yl)propoxy]-6-[(2,4,6-trimethylphenyl)amino]-1,3,5-triazin-2-yl}amino)benzonitrile, HIV-1 reverse transcriptase, p51 subunit, ... | | Authors: | Mislak, A.C, Frey, K.M, Anderson, K.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | A mechanistic and structural investigation of modified derivatives of the diaryltriazine class of NNRTIs targeting HIV-1 reverse transcriptase.
Biochim.Biophys.Acta, 1840, 2014
|
|
4WE1
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ600) | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, Gag-Pol polyprotein, MAGNESIUM ION, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Picomolar Inhibitors of HIV-1 Reverse Transcriptase: Design and Crystallography of Naphthyl Phenyl Ethers.
Acs Med.Chem.Lett., 5, 2014
|
|
3HVT
 
 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | Deposit date: | 1994-07-25 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
5OVN
 
 | | Crystal Structure of FIV Reverse Transcriptase | | Descriptor: | POL protein | | Authors: | Galilee, M, Alian, A. | | Deposit date: | 2017-08-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | The structure of FIV reverse transcriptase and its implications for non-nucleoside inhibitor resistance.
PLoS Pathog., 14, 2018
|
|
2YKM
 
 | | Crystal structure of HIV-1 Reverse Transcriptase (RT) in complex with a Difluoromethylbenzoxazole (DFMB) Pyrimidine Thioether derivative, a non-nucleoside RT inhibitor (NNRTI) | | Descriptor: | 2-[DIFLUORO-[(4-METHYL-PYRIMIDINYL)-THIO]METHYL]-BENZOXAZOLE, CALCIUM ION, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Boyer, J, Arnoult, E, Medebielle, M, Guillemont, J, Unge, T, Unge, J, Jochmans, D. | | Deposit date: | 2011-05-28 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Difluoromethylbenzoxazole Pyrimidine Thioether Derivatives: A Novel Class of Potent Non-Nucleoside HIV-1 Reverse Transcriptase Inhibitors.
J.Med.Chem., 54, 2011
|
|
6KDO
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y/M184V/F160M:DNA:lamivudine 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDK
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
5DMQ
 
 | |
4B3P
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | DNA, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.839 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4B3O
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, 5'-D(*CP*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP *TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3', 5'-R(*AP*UP*GP*AP*3DRP*GP*GP*CP*CP*AP*CP*AP*AP*UP*AP *AP*CP*UP*AP*UP*AP*GP*GP*CP*AP*UP*A)-3', ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4DG1
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with polymorphism mutation K172A and K173A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tu, X, Kirby, K.A, Marchand, B, Sarafianos, S.G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | HIV-1 Reverse Transcriptase (RT) Polymorphism 172K Suppresses the Effect of Clinically Relevant Drug Resistance Mutations to Both Nucleoside and Non-nucleoside RT Inhibitors.
J.Biol.Chem., 287, 2012
|
|
7O24
 
 | |
