1ROU
 
 | | STRUCTURE OF FKBP59-I, THE N-TERMINAL DOMAIN OF A 59 KDA FK506-BINDING PROTEIN, NMR, 22 STRUCTURES | | Descriptor: | FKBP59-I | | Authors: | Craescu, C.T, Rouviere, N, Popescu, A, Cerpolini, E, Lebeau, M.-C, Baulieu, E.-E, Mispelter, J. | | Deposit date: | 1996-06-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the immunophilin-like domain of FKBP59 in solution.
Biochemistry, 35, 1996
|
|
1ROT
 
 | | STRUCTURE OF FKBP59-I, THE N-TERMINAL DOMAIN OF A 59 KDA FK506-BINDING PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FKBP59-I | | Authors: | Craescu, C.T, Rouviere, N, Popescu, A, Cerpolini, E, Lebeau, M.-C, Baulieu, E.-E, Mispelter, J. | | Deposit date: | 1996-06-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the immunophilin-like domain of FKBP59 in solution.
Biochemistry, 35, 1996
|
|
1PBA
 
 | | THE NMR STRUCTURE OF THE ACTIVATION DOMAIN ISOLATED FROM PORCINE PROCARBOXYPEPTIDASE B | | Descriptor: | PROCARBOXYPEPTIDASE B | | Authors: | Vendrell, J, Wider, G, Billeter, M, Aviles, F.X, Wuthrich, K. | | Deposit date: | 1991-11-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the activation domain isolated from porcine procarboxypeptidase B.
EMBO J., 10, 1991
|
|
1OS8
 
 | | RECOMBINANT STREPTOMYCES GRISEUS TRYPSIN | | Descriptor: | CALCIUM ION, SULFATE ION, trypsin | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-18 | | Release date: | 2003-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
1FW6
 
 | | CRYSTAL STRUCTURE OF A TAQ MUTS-DNA-ADP TERNARY COMPLEX | | Descriptor: | 5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3', 5'-D(*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Junop, M.S, Obmolova, G, Rausch, K, Hsieh, P, Yang, W. | | Deposit date: | 2000-09-21 | | Release date: | 2001-02-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair.
Mol.Cell, 7, 2001
|
|
1OSS
 
 | | T190P STREPTOMYCES GRISEUS TRYPSIN IN COMPLEX WITH BENZAMIDINE | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-20 | | Release date: | 2003-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
1ST7
 
 | | Solution structure of Acyl Coenzyme A Binding Protein from yeast | | Descriptor: | Acyl-CoA-binding protein | | Authors: | Teilum, K, Thormann, T, Caterer, N.R, Poulsen, H.I, Jensen, P.H, Knudsen, J, Kragelund, B.B, Poulsen, F.M. | | Deposit date: | 2004-03-25 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Different secondary structure elements as scaffolds for protein folding transition states of two homologous four-helix bundles
Proteins, 59, 2005
|
|
1VCD
 
 | | Crystal Structure of a T.thermophilus HB8 Ap6A hydrolase Ndx1 | | Descriptor: | GLYCEROL, Ndx1, SULFATE ION | | Authors: | Iwai, T, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-05 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Nudix Protein Ndx1 from Thermus thermophilus HB8 in binary complex with diadenosine hexaphosphate
To be Published
|
|
2KPV
 
 | |
1VC9
 
 | | Crystal Structure of a T.thermophilus HB8 Ap6A hydrolase E50Q mutant-Mg2+-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ndx1 | | Authors: | Iwai, T, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-05 | | Release date: | 2005-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Nudix Protein Ndx1 from Thermus thermophilus HB8 in binary complex with diadenosine hexaphosphate
To be Published
|
|
2LKX
 
 | | NMR structure of the homeodomain of Pitx2 in complex with a TAATCC DNA binding site | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3'), Pituitary homeobox 3 | | Authors: | Baird-Titus, J.M, Doerdelmann, T, Chaney, B.A, Clark-Baldwin, K, Dave, V, Ma, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome
Biochemistry, 44, 2005
|
|
2DM5
 
 | | Thermodynamic Penalty Arising From Burial of a Ligand Polar Group Within a Hydrophobic Pocket of a Protein Receptor | | Descriptor: | CADMIUM ION, Major Urinary Protein, OCTANE-1,8-DIOL | | Authors: | Barratt, E, Bronowska, A, Vondrasek, J, Bingham, R, Phillips, S, Homans, S.W. | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic penalty arising from burial of a ligand polar group within a hydrophobic pocket of a protein receptor
J.Mol.Biol., 362, 2006
|
|
2E11
 
 | | The Crystal Structure of XC1258 from Xanthomonas campestris: A CN-hydrolase Superfamily Protein with an Arsenic Adduct in the Active Site | | Descriptor: | CACODYLATE ION, Hydrolase | | Authors: | Chin, K.-H, Tsai, Y.-D, Chan, N.-L, Huang, K.-F, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2006-10-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The crystal structure of XC1258 from Xanthomonas campestris: A putative procaryotic Nit protein with an arsenic adduct in the active site
Proteins, 69, 2007
|
|
1DXR
 
 | | Photosynthetic reaction center from Rhodopseudomonas viridis - His L168 Phe mutant (terbutryn complex) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, 2-T-BUTYLAMINO-4-ETHYLAMINO-6-METHYLTHIO-S-TRIAZINE, BACTERIOCHLOROPHYLL B, ... | | Authors: | Lancaster, C.R.D, Bibikova, M, Sabatino, P, Oesterhelt, D, Michel, H. | | Deposit date: | 2000-01-15 | | Release date: | 2001-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the drastically increased initial electron transfer rate in the reaction center from a Rhodopseudomonas viridis mutant described at 2.00-A resolution.
J. Biol. Chem., 275, 2000
|
|
1EP7
 
 | | CRYSTAL STRUCTURE OF WT THIOREDOXIN H FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | THIOREDOXIN CH1, H-TYPE | | Authors: | Menchise, V, Corbier, C, Didierjean, C, Saviano, M, Benedetti, E, Jacquot, J.P, Aubry, A. | | Deposit date: | 2000-03-28 | | Release date: | 2001-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism.
Biochem.J., 359, 2001
|
|
19HC
 
 | | NINE-HAEM CYTOCHROME C FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ACETATE ION, PROTEIN (NINE-HAEM CYTOCHROME C), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matias, P.M, Coelho, R, Pereira, I.A.C, Coelho, A.V, Thompson, A.W, Sieker, L, Gall, J.L, Carrondo, M.A. | | Deposit date: | 1998-12-01 | | Release date: | 1999-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The primary and three-dimensional structures of a nine-haem cytochrome c from Desulfovibrio desulfuricans ATCC 27774 reveal a new member of the Hmc family.
Structure Fold.Des., 7, 1999
|
|
2CBO
 
 | | Crystal structure of the neocarzinostatin 3Tes24 mutant bound to testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, SULFATE ION, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2CBT
 
 | | Crystal structure of the neocarzinostatin 4Tes1 mutant bound testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2CBM
 
 | | Crystal structure of the apo-form of a neocarzinostatin mutant evolved to bind testosterone. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NEOCARZINOSTATIN | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2CJF
 
 | | TYPE II DEHYDROQUINASE INHIBITOR COMPLEX | | Descriptor: | (1S,4S,5S)-1,4,5-TRIHYDROXY-3-[3-(PHENYLTHIO)PHENYL]CYCLOHEX-2-ENE-1-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Payne, R.J, Riboldi-Tunnicliffe, A, Abell, A.D, Lapthorn, A.J, Abell, C. | | Deposit date: | 2006-03-31 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis, and structural studies on potent biaryl inhibitors of type II dehydroquinases.
Chemmedchem, 2, 2007
|
|
2CJG
 
 | | Lysine aminotransferase from M. tuberculosis in bound PMP form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-LYSINE-EPSILON AMINOTRANSFERASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2006-04-01 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Direct Evidence for a Glutamate Switch Necessary for Substrate Recognition: Crystal Structures of Lysine Epsilon-Aminotransferase (Rv3290C) from Mycobacterium Tuberculosis H37Rv.
J.Mol.Biol., 362, 2006
|
|
2CJD
 
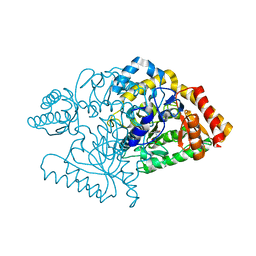 | |
2CIN
 
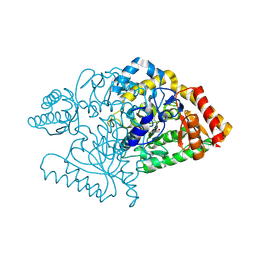 | |
2MJS
 
 | | Anoplin R5K T8W in DPC micelles | | Descriptor: | Anoplin, UNKNOWN ATOM OR ION, dodecyl 2-(trimethylammonio)ethyl phosphate | | Authors: | Uggerhoej, L, Poulsen, T.J, Wimmer, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Alpha-Helical Antimicrobial Peptides: Do's and Don'ts.
Chembiochem, 16, 2015
|
|
1CSR
 
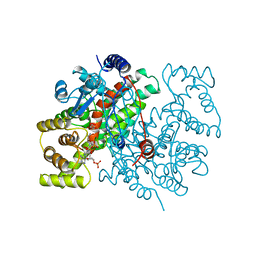 | |
