1ONW
 
 | | Crystal structure of Isoaspartyl Dipeptidase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isoaspartyl dipeptidase, ... | | Authors: | Thoden, J.B, Marti-Arbona, R, Raushel, F.M, Holden, H.M. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High Resolution X-ray Structure of
Isoaspartyl Dipeptidase from
Escherichia coli
Biochemistry, 42, 2003
|
|
1P2H
 
 | | H61M mutant of flavocytochrome c3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALATE LIKE INTERMEDIATE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rothery, E.L, Mowat, C.G, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2003-04-15 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histidine 61: An Important Heme Ligand in the Soluble Fumarate Reductase from Shewanella
frigidimarina
Biochemistry, 42, 2003
|
|
1ORN
 
 | |
3I8W
 
 | |
1OXV
 
 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION, ... | | Authors: | Verdon, G, Albers, S.V, Dijkstra, B.W, Driessen, A.J, Thunnissen, A.M. | | Deposit date: | 2003-04-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus:
nucleotide-free and nucleotide-bound conformations
J.Mol.Biol., 330, 2003
|
|
1P2E
 
 | | H61A mutant of flavocytochrome c3 | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Rothery, E.L, Mowat, C.G, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2003-04-15 | | Release date: | 2003-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 61: An Important Heme Ligand in the Soluble Fumarate Reductase from Shewanella frigidimarina
Biochemistry, 42, 2003
|
|
1OYT
 
 | | COMPLEX OF RECOMBINANT HUMAN THROMBIN WITH A DESIGNED FLUORINATED INHIBITOR | | Descriptor: | (3ASR,4RS,8ASR,8BRS)-4-(2-(4-FLUOROBENZYL)-1,3-DIOXODEACAHYDROPYRROLO[3,4-A] PYRROLIZIN-4-YL)BENZAMIDINE, CALCIUM ION, Hirudin IIB, ... | | Authors: | Banner, D.W, Olsen, J.A. | | Deposit date: | 2003-04-07 | | Release date: | 2003-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Fluorine Scan of Thrombin Inhibitors to Map the Fluorophilicity/Fluorophobicity of an Enzyme Active Site: Evidence for CF...C=O Interactions.
Angew.Chem.Int.Ed.Engl., 42, 2003
|
|
1P0S
 
 | | Crystal Structure of Blood Coagulation Factor Xa in Complex with Ecotin M84R | | Descriptor: | Coagulation factor X precursor, Ecotin precursor, MAGNESIUM ION, ... | | Authors: | Wang, S.X, Hur, E, Sousa, C.A, Brinen, L, Slivka, E.J, Fletterick, R.J. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-26 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Extended Interactions and Gla Domain of Blood Coagulation Factor Xa
Biochemistry, 42, 2003
|
|
1PJ6
 
 | | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with folic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FOLIC ACID, N,N-dimethylglycine oxidase, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
1PJ5
 
 | | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with acetate | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, N,N-dimethylglycine oxidase, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
4E0S
 
 | | Crystal Structure of C5b-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C5, ... | | Authors: | Aleshin, A.E, Stec, B, DiScipio, R, Liddington, R.C. | | Deposit date: | 2012-03-05 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.21 Å) | | Cite: | Crystal structure of c5b-6 suggests structural basis for priming assembly of the membrane attack complex.
J.Biol.Chem., 287, 2012
|
|
4D8S
 
 | | Influenza NA in complex with antiviral compound | | Descriptor: | CALCIUM ION, Neuraminidase, pentan-3-yl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid | | Authors: | Kerry, P.S, Russell, R.J.M.R. | | Deposit date: | 2012-01-11 | | Release date: | 2013-02-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Exploring the interactions of unsaturated glucuronides with influenza virus sialidase.
J.Med.Chem., 55, 2012
|
|
1PJ7
 
 | | Structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with folinic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N,N-dimethylglycine oxidase, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
4D94
 
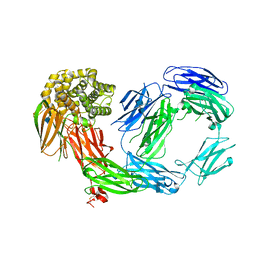 | | Crystal Structure of TEP1r | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Le, B.V, Williams, M, Logarajah, S, Baxter, R.H.G. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis for Genetic Resistance of Anopheles gambiae to Plasmodium: Structural Analysis of TEP1 Susceptible and Resistant Alleles.
Plos Pathog., 8, 2012
|
|
4CZ8
 
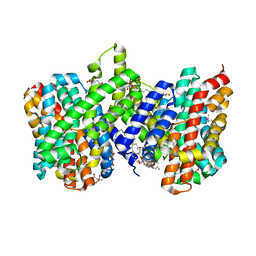 | | Structure of the sodium proton antiporter PaNhaP from Pyrococcus abyssii at pH 8. | | Descriptor: | CITRATE ANION, NA+/H+ ANTIPORTER, PUTATIVE, ... | | Authors: | Woehlert, D, Kuhlbrandt, W, Yildiz, O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure and Substrate Ion Binding in the Sodium/Proton Antiporter Panhap.
Elife, 3, 2014
|
|
4CZA
 
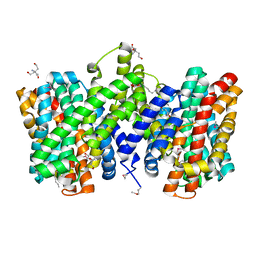 | | Structure of the sodium proton antiporter PaNhaP from Pyrococcus abyssii with bound thallium ion. | | Descriptor: | ACETATE ION, NA+/H+ ANTIPORTER, PUTATIVE, ... | | Authors: | Woehlert, D, Kuhlbrandt, W, Yildiz, O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and substrate ion binding in the sodium/proton antiporter PaNhaP.
Elife, 3, 2014
|
|
1NHX
 
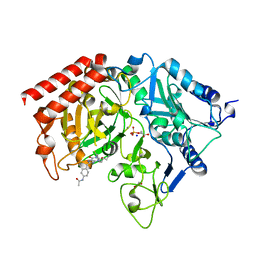 | | PEPCK COMPLEX WITH A GTP-COMPETITIVE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, N-{4-[1-(2-FLUOROBENZYL)-3-BUTYL-2,6-DIOXO-2,3,6,7-TETRAHYDRO-1H-PURIN-8-YLMETHYL]-PHENYL}-ACETAMIDE, ... | | Authors: | Foley, L.H, Wang, P, Dunten, P, Ramsey, G, Gubler, M.-L, Wertheimer, S.J. | | Deposit date: | 2002-12-19 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-RAY STRUCTURES OF TWO XANTHINE INHIBITORS BOUND TO PEPCK and N-3 modifications of substituted 1,8-Dibenzylxanthines
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1PZ0
 
 | |
1Q22
 
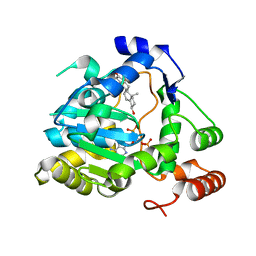 | | Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of DHEA and PAP | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Lee, K.A, Fuda, H, Lee, Y.C, Negishi, M, Strott, C.A, Pedersen, L.C. | | Deposit date: | 2003-07-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone
and 3'-phosphoadenosine 5'-phosphate. Rationale for specificity differences between prototypical
SULT2A1 and the SULT2BG1 isoforms.
J.Biol.Chem., 278, 2003
|
|
3JPW
 
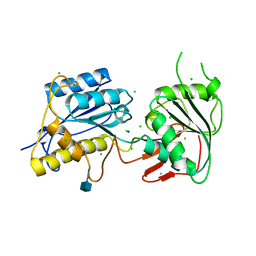 | | Crystal structure of amino terminal domain of the NMDA receptor subunit NR2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutamate [NMDA] receptor subunit epsilon-2, ... | | Authors: | Karakas, E, Simorowski, N, Furukawa, H. | | Deposit date: | 2009-09-04 | | Release date: | 2009-12-08 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit.
Embo J., 28, 2009
|
|
1NT6
 
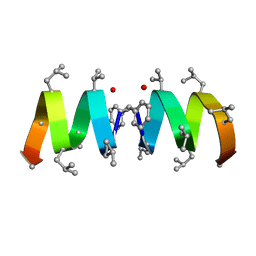 | | F1-Gramicidin C In Sodium Dodecyl Sulfate Micelles (NMR) | | Descriptor: | GRAMICIDIN C | | Authors: | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-11 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
1O1Z
 
 | |
3JU4
 
 | | Crystal Structure Analysis of EndosialidaseNF at 0.98 A Resolution | | Descriptor: | CHLORIDE ION, Endo-N-acetylneuraminidase, N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Neuman, P, Gerardy-Schahn, R, Sheldrick, G.M, Ficner, R. | | Deposit date: | 2009-09-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure analysis of endosialidase NF at 0.98 A resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1N76
 
 | | CRYSTAL STRUCTURE OF HUMAN SEMINAL LACTOFERRIN AT 3.4 A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN, ... | | Authors: | Kumar, J, Weber, W, Munchau, S, Yadav, S, Singh, S.B, Sarvanan, K, Paramsivam, M, Sharma, S, Kaur, P, Bhushan, A, Srinivasan, A, Betzel, C, Singh, T.P. | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of human seminal lactoferrin at 3.4A resolution
Indian J.Biochem.Biophys., 40, 2003
|
|
4ETK
 
 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
