8QBE
 
 | |
7U99
 
 | | EGFR kinase in complex with a macrocyclic inhibitor | | Descriptor: | 19-chloro-22-methoxy-8,9,11,12,14,15-hexahydro-21H-4,6-ethenopyrimido[5,4-m][1,4,7,10,15]benzotetraoxazacycloheptadecine, CITRATE ANION, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Macrocyclization of Quinazoline-Based EGFR Inhibitors Leads to Exclusive Mutant Selectivity for EGFR L858R and Del19.
J.Med.Chem., 65, 2022
|
|
8QB9
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture -PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol 30:20:20:30) | | Descriptor: | Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
7U98
 
 | | EGFR(T790M/V948R) in complex with a macrocyclic inhibitor | | Descriptor: | 19-chloro-18-fluoro-22-methoxy-8,9,11,12,14,15-hexahydro-21H-4,6-ethenopyrimido[5,4-m][1,4,7,10,15]benzotetraoxazacycloheptadecine, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Macrocyclization of Quinazoline-Based EGFR Inhibitors Leads to Exclusive Mutant Selectivity for EGFR L858R and Del19.
J.Med.Chem., 65, 2022
|
|
8KIF
 
 | | The structure of MmaE with substrate | | Descriptor: | (3R)-3-(2-hydroxy-2-oxoethylamino)decanoic acid, FE (II) ION, Putative dioxygenase | | Authors: | Chen, J, Zhou, J. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Variation in biosynthesis and metal-binding properties of isonitrile-containing peptides produced by Mycobacteria versus Streptomyces.
Acs Catalysis, 14, 2024
|
|
8BHH
 
 | | The crystal structure of a feruloyl esterase C from Fusarium oxysporum in complex with p-coumaric acid | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-HYDROXYCINNAMIC ACID, ... | | Authors: | Dimarogona, M, Topakas, E, Kosinas, C, Ferousi, C, Nikolaivits, E. | | Deposit date: | 2022-10-31 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of the Fusarium oxysporum tannase-like feruloyl esterase FaeC in complex with p-coumaric acid provides insight into ligand binding.
Febs Lett., 597, 2023
|
|
6QNK
 
 | | Antibody FAB fragment targeting Gi protein heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, FAB heavy chain, ... | | Authors: | Tsai, C.-J, Muehle, J, Pamula, F, Dawson, R.J.P, Maeda, S, Deupi, X, Schertler, G.F.X. | | Deposit date: | 2019-02-11 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-EM structure of the rhodopsin-G alpha i-beta gamma complex reveals binding of the rhodopsin C-terminal tail to the G beta subunit.
Elife, 8, 2019
|
|
6GQB
 
 | | Cryo-EM reconstruction of yeast 80S ribosome in complex with mRNA, tRNA and eEF2 (GDP+AlF4/sordarin) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pellegrino, S, Yusupov, M, Yusupova, G, Hashem, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-11 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.
J. Mol. Biol., 430, 2018
|
|
5JMQ
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP3 | | Descriptor: | 1,2-ETHANEDIOL, 9-[(5E)-7-carbamimidamido-5,6,7-trideoxy-beta-D-ribo-hept-5-enofuranosyl]-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
Febs J., 284, 2017
|
|
5JAP
 
 | |
6QNJ
 
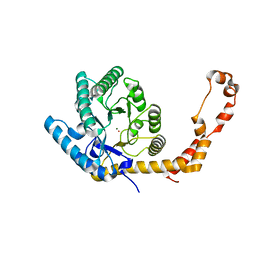 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 4.5 s timepoint | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6ZMB
 
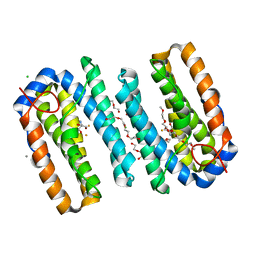 | | Structure of the native tRNA-Monooxygenase enzyme MiaE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Carpentier, P, Atta, M. | | Deposit date: | 2020-07-02 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Nucleic Acids Res., 48, 2020
|
|
8Q6W
 
 | |
5JAT
 
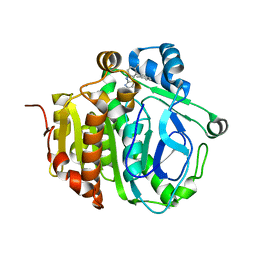 | |
8B4C
 
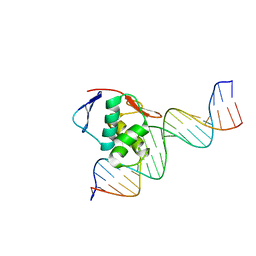 | | ToxR bacterial transcriptional regulator bound to 20 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (20-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8J2F
 
 | | Human neutral shpingomyelinase | | Descriptor: | HEPTANE, MAGNESIUM ION, Sphingomyelin phosphodiesterase 2, ... | | Authors: | Zhang, S.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Molecular basis for the catalytic mechanism of human neutral sphingomyelinases 1 (hSMPD2).
Nat Commun, 14, 2023
|
|
8B4B
 
 | | ToxR bacterial transcriptional regulator bound to 19 bp ompU promoter DNA | | Descriptor: | AMMONIUM ION, CADMIUM ION, Cholera toxin transcriptional activator, ... | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6ZQP
 
 | | Structure of the Pmt2-MIR domain with bound ligands | | Descriptor: | GLYCEROL, PMT2 isoform 1, SULFATE ION, ... | | Authors: | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | Deposit date: | 2020-07-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
8B4D
 
 | | ToxR bacterial transcriptional regulator bound to 40 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (40-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6SRL
 
 | |
5JAR
 
 | |
6GN6
 
 | | Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus | | Descriptor: | Alpha-L-fucosidase, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Kovalova, T, Koval, T, Lipovova, P, Dohnalek, J. | | Deposit date: | 2018-05-30 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active site complementation and hexameric arrangement in the GH family 29; a structure-function study of alpha-l-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus.
Glycobiology, 29, 2019
|
|
8B4U
 
 | | The crystal structure of PET46, a PETase enzyme from Candidatus bathyarchaeota | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Applegate, V, Schumacher, J, Smits, S.H.J. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | An archaeal lid-containing feruloyl esterase degrades polyethylene terephthalate.
Commun Chem, 6, 2023
|
|
6GLW
 
 | | Structure of galectin-10 in complex with the Fab fragment of a Charcot-Leyden crystal solubilizing antibody, 1D11 | | Descriptor: | ACETATE ION, Fab 1D11 - Heavy chain, Fab 1D11 - Light chain, ... | | Authors: | Verstraete, K, Verschueren, K, Savvides, S.N. | | Deposit date: | 2018-05-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein crystallization promotes type 2 immunity and is reversible by antibody treatment.
Science, 364, 2019
|
|
5LYY
 
 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 3-[2-(4-fluoranylphenoxy)ethyl]-1,3-diazaspiro[4.5]decane-2,4-dione, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
