8BJQ
 
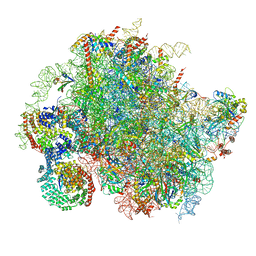 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
6ME2
 
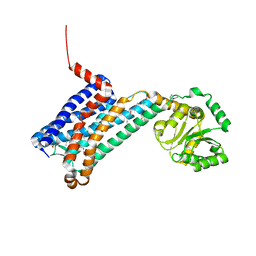 | | XFEL crystal structure of human melatonin receptor MT1 in complex with ramelteon | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
7FYF
 
 | | Crystal Structure of human FABP4 in complex with 4-(4-fluorophenyl)-2-piperidin-1-yl-3-(1H-tetrazol-5-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine, i.e. SMILES c1(ccc(cc1)F)c1c(c(N2CCCCC2)nc2c1COCC2)C1=NN=NN1 with IC50=0.42193 microM | | Descriptor: | (3M)-4-(4-fluorophenyl)-2-(piperidin-1-yl)-3-(1H-tetrazol-5-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FX6
 
 | | Crystal Structure of human FABP4 in complex with N,N-diethyl-4-pyridin-4-yl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridin-2-amine, i.e. SMILES n1c(c(c(c2c1CCCCC2)c1ccncc1)C1=NN=NN1)N(CC)CC with IC50=0.0281951 microM | | Descriptor: | (3M)-N,N-diethyl-4-(pyridin-4-yl)-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridin-2-amine, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
9NBS
 
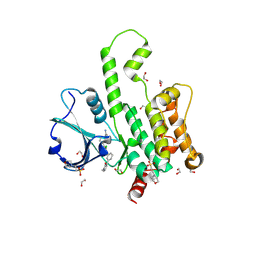 | | Structure of HPK1 with compound R2 | | Descriptor: | (1R,3S,5'P)-4'-chloro-3-methyl-5'-[(3M)-3-(1-methyl-2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1',2'-dihydrospiro[cyclopentane-1,3'-pyrrolo[2,3-b]pyridine]-3-carboxamide, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kiefer, J.R, Walters, B.T, Wang, W, Wu, P, Duo, Y. | | Deposit date: | 2025-02-14 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integrating Hydrogen Exchange with Molecular Dynamics for Improved Ligand Binding Predictions.
J.Chem.Inf.Model., 65, 2025
|
|
1UFI
 
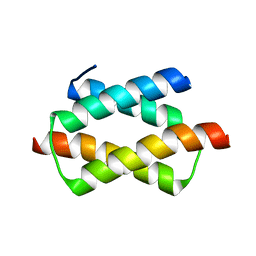 | | Crystal structure of the dimerization domain of human CENP-B | | Descriptor: | Major centromere autoantigen B | | Authors: | Tawaramoto, M.S, Kurumizaka, H, Tanaka, Y, Park, S.-Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-30 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human centromere protein B (CENP-B) dimerization domain at 1.65-A resolution
J.Biol.Chem., 278, 2003
|
|
6TIK
 
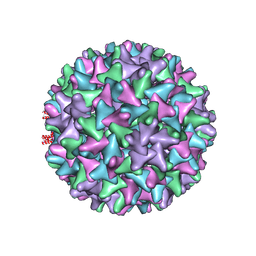 | | Hepatitis B virus core shell--virus-like particle with NadA epitope | | Descriptor: | Capsid protein,Putative adhesin/invasin,Capsid protein,Factor H-binding protein | | Authors: | Roseman, A.M, Colllins, R.F, Derrick, J.P. | | Deposit date: | 2019-11-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An assessment of the use of Hepatitis B Virus core protein virus-like particles to display heterologous antigens from Neisseria meningitidis.
Vaccine, 38, 2020
|
|
6FF7
 
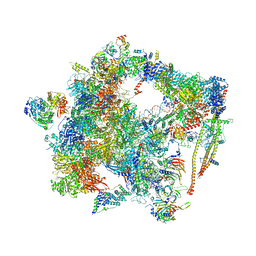 | | human Bact spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, BUD13 homolog, ... | | Authors: | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | Deposit date: | 2018-01-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
8BDR
 
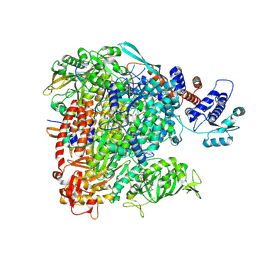 | |
5QIN
 
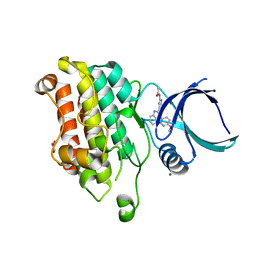 | |
5QIM
 
 | |
5QIL
 
 | |
7G19
 
 | | Crystal Structure of human FABP4 in complex with 5-[2-(1H-tetrazol-5-yl)ethyl]-6,7,8,9,10,11-hexahydrocycloocta[b]indole, i.e. SMILES N1(c2c(C3=C1CCCCCC3)cccc2)CCC1=NN=NN1 with IC50=0.606 microM | | Descriptor: | 5-[2-(1H-tetrazol-5-yl)ethyl]-6,7,8,9,10,11-hexahydro-5H-cycloocta[b]indole, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kyburz, E, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | Descriptor: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
9NA2
 
 | | IRAK4 in Complex with Compound 9 | | Descriptor: | (6P)-6-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-N-[(2R)-2-fluoro-3-hydroxy-3-methylbutyl]-4-(methylamino)pyridine-3-carboxamide, DIMETHYL SULFOXIDE, Interleukin-1 receptor-associated kinase 4 | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 68, 2025
|
|
9NA5
 
 | | IRAK4 in Complex with Compound 24 | | Descriptor: | (6P)-6-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-4-({(1s,4S)-4-[1-(difluoromethyl)-1H-pyrazol-4-yl]cyclohexyl}amino)-N-[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]pyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 68, 2025
|
|
9NA3
 
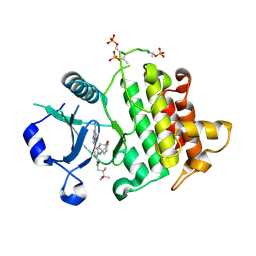 | | IRAK4 in Complex with Compound 15 | | Descriptor: | (6P)-6-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-N-[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]-4-[(4-hydroxybicyclo[2.2.2]octan-1-yl)amino]pyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 68, 2025
|
|
9NA6
 
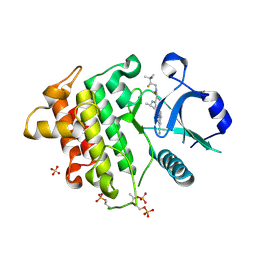 | | IRAK4 in Complex with Compound 34 | | Descriptor: | (6P)-4-{[(1S)-1-cyanoethyl]amino}-6-[(8S)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-N-[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]pyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 68, 2025
|
|
6KRL
 
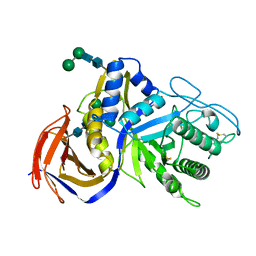 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Substrate recognition by a bifunctional GH30-7 xylanase B from Talaromyces cellulolyticus.
Febs Open Bio, 10, 2020
|
|
9NA4
 
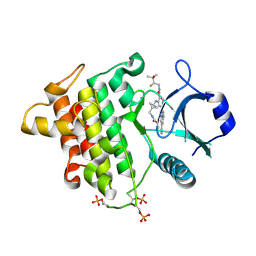 | | IRAK4 in Complex with Compound 18 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, methyl {[(1S,3s)-3-{[(2P)-2-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-5-{[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]carbamoyl}pyridin-4-yl]amino}cyclobutyl]methyl}carbamate | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 68, 2025
|
|
7OB1
 
 | | OLIGOPEPTIDASE B FROM S. PROTEOMACULANS WITH MODIFIED HINGE | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Korzhenevskiy, D.A, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
7P0Z
 
 | | 2.43 A Mycobacterium marinum EspB. | | Descriptor: | ESX-1 secretion-associated protein EspB | | Authors: | Gijsbers, A, Zhang, Y, Vinciauskaite, V, Siroy, A, Ye, G, Tria, G, Mathew, A, Sanchez-Puig, N, Lopez-Iglesias, C, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Priming mycobacterial ESX-secreted protein B to form a channel-like structure.
Curr Res Struct Biol, 3, 2021
|
|
7P13
 
 | | 2.29 A Mycobacterium tuberculosis EspB. | | Descriptor: | ESX-1 secretion-associated protein EspB | | Authors: | Gijsbers, A, Zhang, Y, Vinciauskaite, V, Siroy, A, Gao, Y, Tria, G, Mathew, A, Sanchez-Puig, N, Lopez-Iglesias, C, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Priming mycobacterial ESX-secreted protein B to form a channel-like structure.
Curr Res Struct Biol, 3, 2021
|
|
8PQU
 
 | |
8BYN
 
 | | Chronic traumatic encephalopathy tau filaments with PET ligand flortaucipir | | Descriptor: | 7-(6-fluoranylpyridin-3-yl)-5~{H}-pyrido[4,3-b]indole, Microtubule-associated protein tau | | Authors: | Shi, Y, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM Structures of Chronic Traumatic Encephalopathy Tau Filaments with PET Ligand Flortaucipir.
J.Mol.Biol., 435, 2023
|
|
