4CDU
 
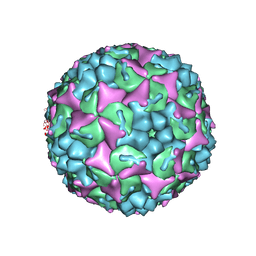 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
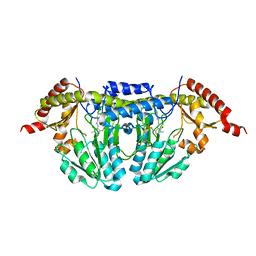
1P3W
 
 | |
3CII
 
 | | Structure of NKG2A/CD94 bound to HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen peptide, HLA class I histocompatibility antigen, ... | | Authors: | Strong, R.K, Kaiser, B.K, Pizarro, J.C. | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.41 Å) | | Cite: | Structural basis for NKG2A/CD94 recognition of HLA-E.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1THD
 
 | | COMPLEX ORGANIZATION OF DENGUE VIRUS E PROTEIN AS REVEALED BY 9.5 ANGSTROM CRYO-EM RECONSTRUCTION | | Descriptor: | Major envelope protein E | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
1T3A
 
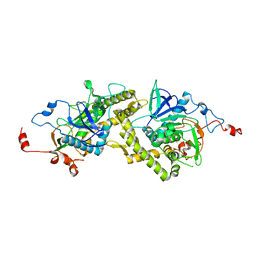 | | Crystal structure of Clostridium botulinum neurotoxin type E catalytic domain | | Descriptor: | CHLORIDE ION, ZINC ION, neurotoxin type E | | Authors: | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2004-04-26 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
3WHI
 
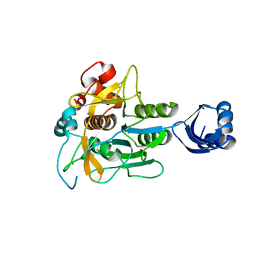 | | Crystal structure of unautoprocessed form of IS1-inserted Pro-subtilisin E | | Descriptor: | CALCIUM ION, Subtilisin E | | Authors: | Uehara, R, Angkawidjaja, C, Koga, Y, Kanaya, S. | | Deposit date: | 2013-08-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Formation of the High-Affinity Calcium Binding Site in Pro-subtilisin E with the Insertion Sequence IS1 of Pro-Tk-subtilisin
Biochemistry, 52, 2013
|
|
3D3X
 
 | |
1TOK
 
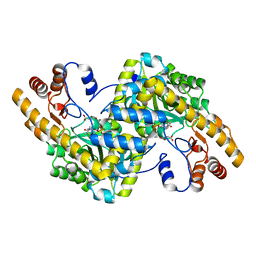 | | Maleic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, MALEIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
4EFA
 
 | | Crystal Structure of the Heterotrimeric EGChead Peripheral Stalk Complex of the Yeast Vacuolar ATPase - second conformation | | Descriptor: | SULFATE ION, V-type proton ATPase subunit C, V-type proton ATPase subunit E, ... | | Authors: | Oot, R.A, Huang, L.S, Berry, E.A, Wilkens, S. | | Deposit date: | 2012-03-29 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8163 Å) | | Cite: | Crystal Structure of the Yeast Vacuolar ATPase Heterotrimeric EGC(head) Peripheral Stalk Complex.
Structure, 20, 2012
|
|
4QMO
 
 | | MST3 IN COMPLEX WITH Imidazolo-oxindole PKR inhibitor C16 | | Descriptor: | (8Z)-8-(1H-imidazol-5-ylmethylidene)-6,8-dihydro-7H-[1,3]thiazolo[5,4-e]indol-7-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
5NAL
 
 | | The crystal structure of inhibitor-15 covalently bound to PDE6D | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ~{N}4-[(4-chlorophenyl)methyl]-~{N}1-(cyclohexylmethyl)-~{N}4-cyclopentyl-~{N}1-[(~{Z})-4-[(~{E})-methyliminomethyl]-5-oxidanyl-hex-4-enyl]benzene-1,4-disulfonamide | | Authors: | Fansa, E.K, Martin-Gago, P, Waldmann, H, Wittinghofer, A. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Covalent Protein Labeling at Glutamic Acids.
Cell Chem Biol, 24, 2017
|
|
2O99
 
 | | The crystal structure of E.coli IclR C-terminal fragment in complex with glyoxylate | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, GLYCOLIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
2OSX
 
 | | Endo-glycoceramidase II from Rhodococcus sp.: Ganglioside GM3 Complex | | Descriptor: | Endoglycoceramidase II, GLYCEROL, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-06 | | Release date: | 2007-02-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and Mechanistic Analyses of endo-Glycoceramidase II, a Membrane-associated Family 5 Glycosidase in the Apo and GM3 Ganglioside-bound Forms.
J.Biol.Chem., 282, 2007
|
|
1PTM
 
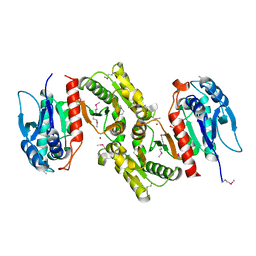 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, PHOSPHATE ION, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-06-23 | | Release date: | 2003-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
4GX8
 
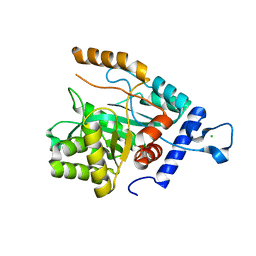 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | CHLORIDE ION, DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Robinson, A, Horan, N, Xu, Z.-Q, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
4DKP
 
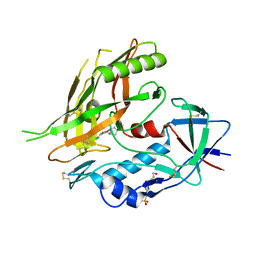 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with AWS-I-50 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(1S,2S)-2-amino-2,3-dihydro-1H-inden-1-yl]-N'-(4-chloro-3-fluorophenyl)ethanediamide, ... | | Authors: | Kwon, Y.D, LaLonde, J.M, Jones, D.M, Sun, A.W, Courter, J.R, Soeta, T, Kobayashi, T, Princiotto, A.M, Wu, X, Mascola, J, Schon, A, Freire, E, Sodroski, J, Madani, N, Smith III, A.B, Kwong, P.D. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7978 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors.
J.Med.Chem., 55, 2012
|
|
2BTD
 
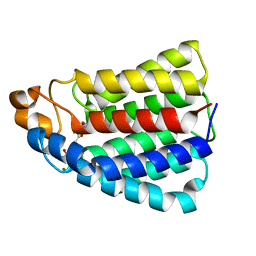 | | Crystal structure of DhaL from E. coli | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PTS-DEPENDENT DIHYDROXYACETONE KINASE | | Authors: | Oberholzer, A.E, Schneider, P, Bachler, C, Baumann, U, Erni, B. | | Deposit date: | 2005-05-27 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Nucleotide-Binding Subunit Dhal of the Escherichia Coli Dihydroxyacetone Kinase.
J.Mol.Biol., 359, 2006
|
|
2O9A
 
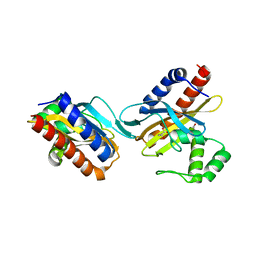 | | The crystal structure of the E.coli IclR C-terminal fragment in complex with pyruvate. | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, PYRUVIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
1JL1
 
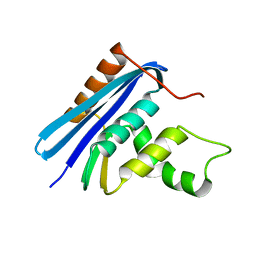 | | D10A E. coli ribonuclease HI | | Descriptor: | RIBONUCLEASE HI | | Authors: | Goedken, E.R, Marqusee, S. | | Deposit date: | 2001-07-13 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Native-state energetics of a thermostabilized variant of ribonuclease HI.
J.Mol.Biol., 314, 2001
|
|
4RZB
 
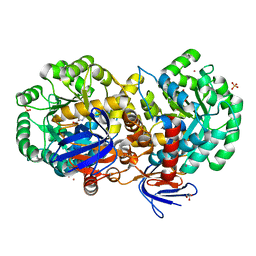 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate, SOAKED WITH MERCURY | | Descriptor: | GLYCEROL, MERCURY (II) ION, N-[(E)-iminomethyl]-L-aspartic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-12-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
3BGR
 
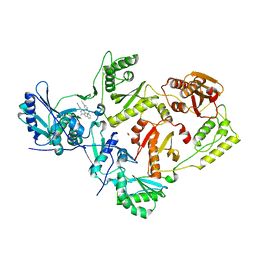 | | Crystal structure of K103N/Y181C mutant HIV-1 reverse transcriptase (RT) in complex with TMC278 (Rilpivirine), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, ... | | Authors: | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | Deposit date: | 2007-11-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3TH8
 
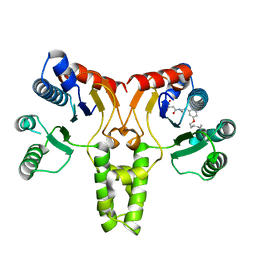 | | Structure of E. coli undecaprenyl diphosphate synthase complexed with BPH-1063 | | Descriptor: | (2Z)-4-({3-[3-(hexyloxy)phenyl]propyl}amino)-2-hydroxy-4-oxobut-2-enoic acid, Undecaprenyl pyrophosphate synthase | | Authors: | Cao, R, Zhu, W, Zhang, Y, Oldfield, E. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
1SFE
 
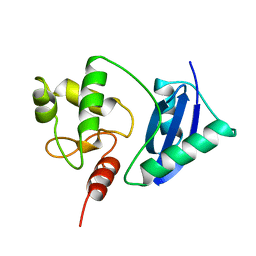 | | ADA O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | ADA O6-METHYLGUANINE-DNA METHYLTRANSFERASE | | Authors: | Moore, M.H, Gulbis, J.M, Dodson, E.J, Demple, B, Moody, P.C.E. | | Deposit date: | 1996-06-21 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a suicidal DNA repair protein: the Ada O6-methylguanine-DNA methyltransferase from E. coli.
EMBO J., 13, 1994
|
|
4RDV
 
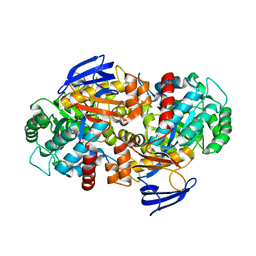 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-FORMIMINO-L-GLUTAMATE IMINOHYDROLASE, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate
To be Published
|
|
1CIS
 
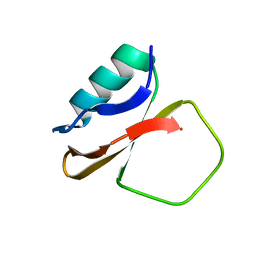 | |
