3KLA
 
 | | Ca2+ release from the endoplasmic reticulum of NY-ESO-1 specific T cells is modulated by the affinity of T cell receptor and by the use of the CD8 co-receptor | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Chen, J.L, Morgan, A.J, Stewart-Jones, G, Shepherd, D, Bossi, G, Wooldridge, L. | | Deposit date: | 2009-11-07 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ca2+ Release from the Endoplasmic Reticulum of NY-ESO-1-Specific T Cells Is Modulated by the Affinity of TCR and by the Use of the CD8 Coreceptor.
J.Immunol., 184, 2010
|
|
7R4H
 
 | | phospho-STING binding to adaptor protein complex-1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Xu, P, Ablasser, A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-09-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Clathrin-associated AP-1 controls termination of STING signalling.
Nature, 610, 2022
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
6ST4
 
 | |
6ADM
 
 | |
9F6Y
 
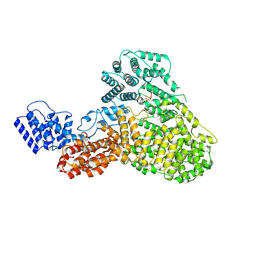 | | CryoEM structure of Human Mediator subunit MED23 complexed with phosphorylated Elk-1 transcription factor | | Descriptor: | Green fluorescent protein,ETS domain-containing protein Elk-1, Mediator of RNA polymerase II transcription subunit 23 | | Authors: | Monte, D, Verger, A, Lens, Z, villeret, V. | | Deposit date: | 2024-05-02 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of human Mediator recruitment by the phosphorylated transcription factor Elk-1.
Nat Commun, 16, 2025
|
|
7WRU
 
 | |
4MSH
 
 | | Crystal Structure of PDE10A2 with fragment ZT0143 ((2S)-4-chloro-2,3-dihydro-1,3-benzothiazol-2-amine) | | Descriptor: | 4-chloro-1,3-benzothiazol-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
9BGF
 
 | |
6ADL
 
 | |
6ADR
 
 | |
9FAZ
 
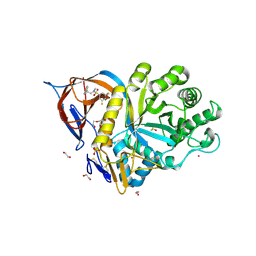 | | Gcase in complex with small molecule inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tisi, D, Cleasby, A. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Fragment-Based Discovery of a Series of Allosteric-Binding Site Modulators of beta-Glucocerebrosidase.
J.Med.Chem., 67, 2024
|
|
5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Kovalevsky, A.Y, Gerlits, O.O. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7LRY
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA, (-)FTC-TP, and CA(2+) ion | | Descriptor: | AMMONIUM ION, CALCIUM ION, DNA/RNA (38-MER), ... | | Authors: | Hoang, A, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.
Drug Discov Today, 27, 2022
|
|
2R5D
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HIV entry inhibitor PIE7, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6H6Q
 
 | | Fragment Derived XIAP inhibitor | | Descriptor: | 2-[(2~{R},5~{R})-2-[[(3~{R},5~{R})-3,5-dimethylmorpholin-4-yl]methyl]-5-methyl-piperazin-1-yl]-1-[6-[(~{S})-(4-fluorophenyl)-oxidanyl-methyl]-3,3-dimethyl-2~{H}-pyrrolo[3,2-b]pyridin-1-yl]ethanone, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Williams, P.A. | | Deposit date: | 2018-07-30 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | A Fragment-Derived Clinical Candidate for Antagonism of X-Linked and Cellular Inhibitor of Apoptosis Proteins: 1-(6-[(4-Fluorophenyl)methyl]-5-(hydroxymethyl)-3,3-dimethyl-1 H,2 H,3 H-pyrrolo[3,2- b]pyridin-1-yl)-2-[(2 R,5 R)-5-methyl-2-([(3R)-3-methylmorpholin-4-yl]methyl)piperazin-1-yl]ethan-1-one (ASTX660).
J. Med. Chem., 61, 2018
|
|
5KAN
 
 | | Crystal structure of multidonor HV1-18-class broadly neutralizing Influenza A antibody 16.g.07 in complex with A/Hong Kong/1-4-MA21-1/1968 (H3N2) Hemagglutinin | | Descriptor: | 16.g.07 Heavy chain, 16.g.07 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell(Cambridge,Mass.), 166, 2016
|
|
7LRX
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA, L-dCTP, and CA(2+) ion | | Descriptor: | 4-amino-1-{2-deoxy-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-L-erythro-pentofuranosyl}pyrimidin-2(1H)-one, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Hoang, A, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.
Drug Discov Today, 27, 2022
|
|
6A1P
 
 | | Mandelate oxidase mutant-Y128F with 5-deazariboflavin mononucleotide and phenylpyruvic acid | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 3-PHENYLPYRUVIC ACID, 4-hydroxymandelate oxidase, ... | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5DQL
 
 | |
3KQ7
 
 | |
9C72
 
 | | Structure of the ASH3 domain of Drosophila melanogaster Spd-2 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Spindle defective 2 | | Authors: | Johnson, S, Feng, Z, Lea, S.M, Raff, J.W. | | Deposit date: | 2024-06-10 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The conserved Spd-2/CEP192 domain adopts a unique protein fold to promote centrosome scaffold assembly.
Sci Adv, 11, 2025
|
|
3WTK
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Thiacloprid | | Descriptor: | Acetylcholine-binding protein, {(2Z)-3-[(6-chloropyridin-3-yl)methyl]-1,3-thiazolidin-2-ylidene}cyanamide | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
7AQF
 
 | |
7QHD
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((2-(1H-indol-3-yl)ethyl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-[2-(1~{H}-indol-3-yl)ethylcarbamoyl]phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
