5UGS
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT501 | | Descriptor: | 5-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]-2-(2-methylphenoxy)phenol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5UHN
 
 | |
4M18
 
 | | Crystal Structure of Surfactant Protein-D D325A/R343V mutant in complex with trimannose (Man-a1,2Man-a1,2Man) | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Goh, B.C, Rynkiewicz, M.J, Cafarella, T.R, White, M.R, Hartshorn, K.L, Allen, K, Crouch, E.C, Calin, O, Seeberger, P.H, Schulten, K, Seaton, B.A. | | Deposit date: | 2013-08-02 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | Molecular mechanisms of inhibition of influenza by surfactant protein d revealed by large-scale molecular dynamics simulation.
Biochemistry, 52, 2013
|
|
4M20
 
 | |
5U14
 
 | | E. coli dihydropteroate synthase complexed with an 8-mercaptoguanine derivative: 4-{2-[(2-amino-6-oxo-6,9-dihydro-1H-purin-8-yl)sulfanyl]ethyl}benzene-1-sulfonamide | | Descriptor: | 4-{2-[(2-amino-6-oxo-6,9-dihydro-1H-purin-8-yl)sulfanyl]ethyl}benzene-1-sulfonamide, Dihydropteroate synthase | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | 8-Mercaptoguanine Derivatives as Inhibitors of Dihydropteroate Synthase.
Chemistry, 24, 2018
|
|
4ECC
 
 | | Chimeric GST Containing Inserts of Kininogen Peptides | | Descriptor: | chimeric protein between GSHKT10 and domain 5 of kininogen-1 | | Authors: | Amber, A.B, Sergei, M.M, Yi, P, Rita, R, Xiaoping, Q, Marianne, P.-C, William, C.M, Vivien, Y, Keith, R.M, Anton, A.K. | | Deposit date: | 2012-03-26 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chimeric glutathione S-transferases containing inserts of kininogen peptides: potential novel protein therapeutics.
J.Biol.Chem., 287, 2012
|
|
5UFP
 
 | | Crystal structure of PT2399 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-({(1S)-7-[(difluoromethyl)sulfonyl]-2,2-difluoro-1-hydroxy-2,3-dihydro-1H-inden-4-yl}oxy)-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2017-01-05 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On-target efficacy of a HIF-2 alpha antagonist in preclinical kidney cancer models.
Nature, 539, 2016
|
|
4LTA
 
 | |
4ES0
 
 | | X-ray structure of WDR5-SETd1b Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase SETD1B, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.817 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
5UGU
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT506 | | Descriptor: | 2-[4-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5UGX
 
 | |
3HIF
 
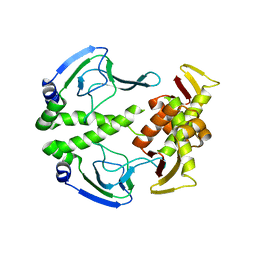 | | The crystal structure of apo wild type CAP at 3.6 A resolution. | | Descriptor: | Catabolite gene activator | | Authors: | Steitz, T.A, Sharma, H, Wang, J, Kong, J, Yu, S. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5UGT
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT504 | | Descriptor: | 2-(2-chloranylphenoxy)-5-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
4E6S
 
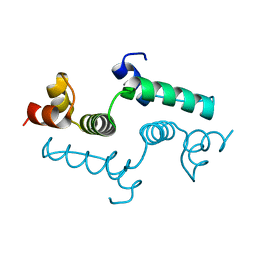 | | Crystal structure of the SCAN domain from mouse Zfp206 | | Descriptor: | Zinc finger and SCAN domain-containing protein 10 | | Authors: | Liang, Y, Choo, S.H, Rossbach, M, Baburajendran, N, Palasingam, P, Kolatkar, P.R. | | Deposit date: | 2012-03-15 | | Release date: | 2012-05-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal optimization and preliminary diffraction data analysis of the SCAN domain of Zfp206.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5UGL
 
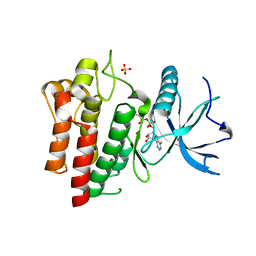 | |
1IDP
 
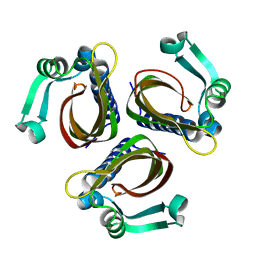 | |
3KRJ
 
 | | cFMS tyrosine kinase in complex with 4-Cyano-1H-imidazole-2-carboxylic acid (2-cyclohex-1-enyl-4-piperidin-4-yl-phenyl)-amide | | Descriptor: | 4-cyano-N-(2-cyclohex-1-en-1-yl-4-piperidin-4-ylphenyl)-1H-imidazole-2-carboxamide, ACETATE ION, Macrophage colony-stimulating factor 1 receptor, ... | | Authors: | Schubert, C. | | Deposit date: | 2009-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of a Potent Class of Arylamide Colony-Stimulating Factor-1 Receptor Inhibitors Leading to Anti-inflammatory Clinical Candidate 4-Cyano-N-[2-(1-cyclohexen-1-yl)-4-[1-[(dimethylamino)acetyl]-4-piperidinyl]phenyl]-1H-imidazole-2-carboxamide (JNJ-28312141).
J.Med.Chem., 54, 2011
|
|
5UKN
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522UCA Fab | | Descriptor: | CHLORIDE ION, DH522UCA Fab fragment heavy chain, DH522UCA Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
5UKP
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522.1 Fab | | Descriptor: | DH522.1 Fab fragment heavy chain, DH522.1 Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
4LST
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC01 in complex with HIV-1 clade C strain ZM176.66 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, HEAVY CHAIN OF ANTIBODY VRC01, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
4EQ5
 
 | | DNA ligase from the archaeon Thermococcus sibiricus | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase | | Authors: | Petrova, T, Bezsudnova, E.Y, Dorokhov, B.D, Slutskaya, E.S, Polyakov, K.M, Dorovatovskiy, P.V, Ravin, N.V, Skryabin, K.G, Kovalchuk, M.V, Popov, V.O. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Expression, purification, crystallization and preliminary crystallographic analysis of a thermostable DNA ligase from the archaeon Thermococcus sibiricus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3HJZ
 
 | | The structure of an aldolase from Prochlorococcus marinus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5UKR
 
 | |
4ERY
 
 | | X-ray structure of WDR5-MLL3 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
5UKO
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522IA Fab | | Descriptor: | DH522IA Fab fragment heavy chain, DH522IA Fab fragment light chain | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
