5ESJ
 
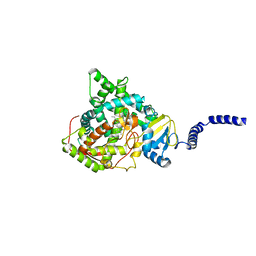 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) G464S mutant complexed with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Impact of Homologous Resistance Mutations from Pathogenic Yeast on Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 62, 2018
|
|
3BYH
 
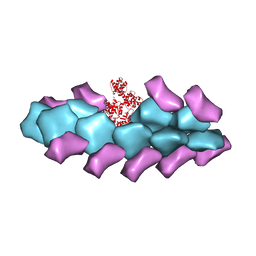 | | Model of actin-fimbrin ABD2 complex | | Descriptor: | Actin, fimbrin ABD2 | | Authors: | Galkin, V.E, Orlova, A, Cherepanova, O, Lebart, M.C, Egelman, E.H. | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1ADC
 
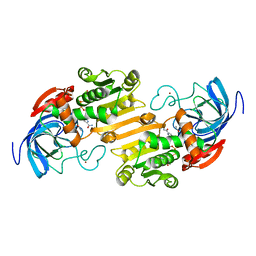 | | CRYSTALLOGRAPHIC STUDIES OF ISOSTERIC NAD ANALOGUES BOUND TO ALCOHOL DEHYDROGENASE: SPECIFICITY AND SUBSTRATE BINDING IN TWO TERNARY COMPLEXES | | Descriptor: | 5-BETA-D-RIBOFURANOSYLPICOLINAMIDE ADENINE-DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, ETHANOL, ... | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-12-13 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
Biochemistry, 33, 1994
|
|
4OIL
 
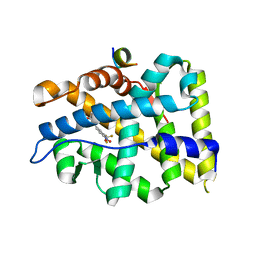 | | Crystal structure of T877A-AR-LBD bound with co-regulator peptide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-19 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
1T9C
 
 | | Crystal Structure Of Yeast Acetohydroxyacid Synthase In Complex With A Sulfonylurea Herbicide, Sulfometuron methyl | | Descriptor: | Acetolactate synthase, mitochondrial, ETHYL DIHYDROGEN DIPHOSPHATE, ... | | Authors: | McCourt, J.A, Pang, S.S, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2004-05-16 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Elucidating the specificity of binding of sulfonylurea herbicides to acetohydroxyacid synthase.
Biochemistry, 44, 2005
|
|
4LVD
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-nitrophenyl)cyclopropanecarboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2WOJ
 
 | | ADP-AlF4 complex of S. cerevisiae GET3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPASE GET3, MAGNESIUM ION, ... | | Authors: | Mateja, A, Szlachcic, A, Downing, M.E, Dobosz, M, Mariappan, M, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2009-07-26 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | The Structural Basis of Tail-Anchored Membrane Protein Recognition by Get3.
Nature, 461, 2009
|
|
7WVR
 
 | |
1D46
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX:-100 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
4OQO
 
 | | Crystal structure of the tryptic generated iron-free C-lobe of bovine Lactoferrin at 2.42 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lactotransferrin | | Authors: | Singh, A, Rastogi, N, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of the iron-free true C-terminal half of bovine lactoferrin produced by tryptic digestion and its functional significance in the gut.
Febs J., 281, 2014
|
|
6KN4
 
 | |
5AG5
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2R)-3-benzyl-2-(1H-indazol-5-yl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
1BTV
 
 | | STRUCTURE OF BET V 1, NMR, 20 STRUCTURES | | Descriptor: | BET V 1 | | Authors: | Osmark, P, Poulsen, F.M, Gajhede, M, Larsen, J.N, Spangfort, M.D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
5AG6
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2R)-2-(4-hydroxy-3-methoxyphenyl)-3-(pyridin-2-ylmethyl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
2W00
 
 | | Crystal structure of the HsdR subunit of the EcoR124I restriction enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HSDR, MAGNESIUM ION | | Authors: | Lapkouski, M, Panjikar, S, Kuta Smatanova, I, Ettrich, R, Csefalvay, E. | | Deposit date: | 2008-08-08 | | Release date: | 2008-12-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Motor Subunit of Type I Restriction-Modification Complex Ecor124I.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2Q72
 
 | | Crystal Structure Analysis of LeuT complexed with L-leucine, sodium, and imipramine | | Descriptor: | 3-(5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, LEUCINE, SODIUM ION, ... | | Authors: | Singh, S.K, Yamashita, A, Gouaux, E. | | Deposit date: | 2007-06-05 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antidepressant binding site in a bacterial homologue of neurotransmitter transporters.
Nature, 448, 2007
|
|
2Q8G
 
 | | Structure of pyruvate dehydrogenase kinase isoform 1 in complex with glucose-lowering drug AZD7545 | | Descriptor: | 4-[(3-CHLORO-4-{[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]AMINO}PHENYL)SULFONYL]-N,N-DIMETHYLBENZAMIDE, POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
6H8Q
 
 | | Structural basis for Scc3-dependent cohesin recruitment to chromatin | | Descriptor: | Cohesin subunit SCC3, DNA (5'-D(P*CP*TP*TP*TP*CP*GP*TP*TP*TP*CP*CP*TP*TP*GP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*CP*AP*AP*GP*GP*AP*AP*AP*CP*GP*AP*AP*AP*G)-3'), ... | | Authors: | Li, Y, Muir, K, Panne, D. | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.631 Å) | | Cite: | Structural basis for Scc3-dependent cohesin recruitment to chromatin.
Elife, 7, 2018
|
|
3BZG
 
 | | UVDE pH4.4 | | Descriptor: | UV endonuclease | | Authors: | Meulenbroek, E.M, Paspaleva, K, Thomassen, E.A.J, Abrahams, J.P, Goosen, N, Pannu, N.S. | | Deposit date: | 2008-01-18 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Involvement of a carboxylated lysine in UV damage endonuclease
Protein Sci., 18, 2009
|
|
4YZZ
 
 | | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site
To be published
|
|
7UUP
 
 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-04-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
2VS2
 
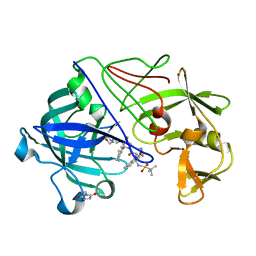 | | Neutron diffraction structure of endothiapepsin in complex with a gem- diol inhibitor. | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Coates, L, Tuan, H.-F, Tomanicek, S, Kovalevsky, A, Mustyakimov, M, Erskine, P, Cooper, J. | | Deposit date: | 2008-04-17 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | The Catalytic Mechanism of an Aspartic Proteinase Explored with Neutron and X-Ray Diffraction
J.Am.Chem.Soc., 130, 2008
|
|
5B77
 
 | |
4MT8
 
 | |
1AQB
 
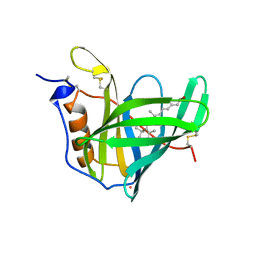 | | RETINOL-BINDING PROTEIN (RBP) FROM PIG PLASMA | | Descriptor: | CADMIUM ION, RETINOL, RETINOL-BINDING PROTEIN | | Authors: | Zanotti, G, Panzalorto, M, Marcato, A, Malpeli, G, Folli, C, Berni, R. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of pig plasma retinol-binding protein at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
