5HWW
 
 | | Crystal structure of PAS1 complexed with 1,2,4-TMB | | Descriptor: | 1,2,4-trimethylbenzene, Sensor histidine kinase TodS | | Authors: | Hwang, J, Koh, S. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Insights into Toluene Sensing in the TodS/TodT Signal Transduction System.
J. Biol. Chem., 291, 2016
|
|
6HQX
 
 | | Human Carbonic Anhydrase II in complex with 4-Ethylbenzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-ethylbenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
4U67
 
 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 | | Descriptor: | 23s RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-07-28 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
1GA9
 
 | | CRYSTAL STRUCTURE OF AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH NON-BETA-LACTAMASE INHIBITOR (2, 3-(4-BENZENESULFONYL-THIOPHENE-2-SULFONYLAMINO)-PHENYLBORONIC ACID) | | Descriptor: | 3-(4-BENZENESULFONYL-THIOPHENE-2-SULFONYLAMINO)-PHENYLBORONIC ACID, BETA-LACTAMASE, PHOSPHATE ION, ... | | Authors: | Tondi, D, Powers, R.A, Caselli, E, Negri, M.C, Blazquez, J, Costi, M.P, Shoichet, B.K. | | Deposit date: | 2000-11-29 | | Release date: | 2001-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design and in-parallel synthesis of inhibitors of AmpC beta-lactamase.
Chem.Biol., 8, 2001
|
|
1QVI
 
 | | Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 Angstrom resolution: flexibility and function in the head | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gourinath, S, Himmel, D.M, Brown, J.H, Reshetnikova, L, Szent-Gyrgyi, A.G, Cohen, C. | | Deposit date: | 2003-08-27 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of scallop Myosin s1 in the pre-power stroke state to 2.6 a resolution: flexibility and function in the head.
Structure, 11, 2003
|
|
8BRW
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13C,I297C | | Descriptor: | Methionine--tRNA ligase, ZINC ION | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8BRX
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13C,I297C complexed with beta-3-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, Methionine--tRNA ligase, ... | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8BRV
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13M,I297C complexed with beta3-methionine. | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, Methionine--tRNA ligase, ... | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
3O32
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with 3,5-dichlorocatechol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, 3,5-dichlorobenzene-1,2-diol, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomytseva, M, Golovleva, L. | | Deposit date: | 2010-07-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
4HT3
 
 | | The crystal structure of Salmonella typhimurium Tryptophan Synthase at 1.30A complexed with N-(4'-TRIFLUOROMETHOXYBENZENESULFONYL)-2-AMINO-1-ETHYLPHOSPHATE (F9) inhibitor in the alpha site, internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-31 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
3O4M
 
 | | Crystal structure of porcine pancreatic phospholipase A2 in complex with 1,2-dihydroxybenzene | | Descriptor: | CALCIUM ION, CATECHOL, Phospholipase A2, ... | | Authors: | Dileep, K.V, Tintu, I, Karthe, P, Mandal, P.K, Haridas, M, Sadasivan, C. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding to PLA(2) may contribute to the anti-inflammatory activity of catechol
Chem.Biol.Drug Des., 2011
|
|
8BRU
 
 | | Escherichia coli methionyl-tRNA synthetase mutant L13M,I297C | | Descriptor: | CITRIC ACID, Methionine--tRNA ligase, ZINC ION | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
2OTG
 
 | | Rigor-like structures of muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2007-02-08 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Rigor-like Structures from Muscle Myosins Reveal Key Mechanical Elements in the Transduction Pathways of This Allosteric Motor.
Structure, 15, 2007
|
|
3O6J
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with hydroxyquinol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, CHLORIDE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
8W07
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-402 | | Descriptor: | (1R,2S,4R)-N-cyclohexyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-cyclohexyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.K, Nettles, K.W. | | Deposit date: | 2024-02-13 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2EC6
 
 | | Placopecten Striated Muscle Myosin II | | Descriptor: | CALCIUM ION, Myosin essential light chain, Myosin heavy chain, ... | | Authors: | Yang, Y, Brown, J, Samudrala, G, Reutzel, R, Szent-Gyorgyi, A. | | Deposit date: | 2007-02-10 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
2QH6
 
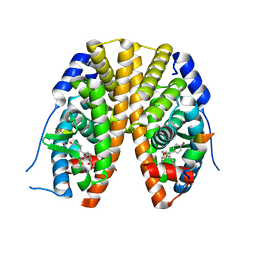 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Complexed with an Oxabicyclic diarylethylene Compound | | Descriptor: | DIETHYL (1R,2S,3R,4S)-5,6-BIS(4-HYDROXYPHENYL)-7-OXABICYCLO[2.2.1]HEPT-5-ENE-2,3-DICARBOXYLATE, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-06-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
2QXM
 
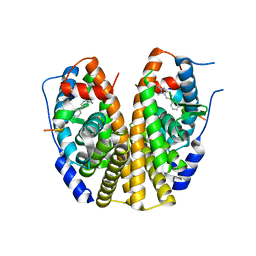 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Complexed to Burned Meat Compound PhIP | | Descriptor: | 2-AMINO-1-METHYL-6-PHENYLIMIDAZO[4,5-B]PYRIDINE, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Gil, G, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-08-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
2QGT
 
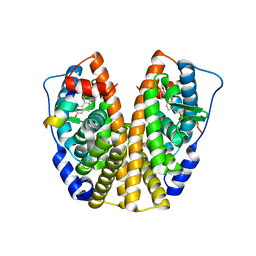 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Complexed to an Ether Estradiol Compound | | Descriptor: | (9BETA,11ALPHA,13ALPHA,14BETA,17ALPHA)-11-(METHOXYMETHYL)ESTRA-1(10),2,4-TRIENE-3,17-DIOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-06-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
3G4Y
 
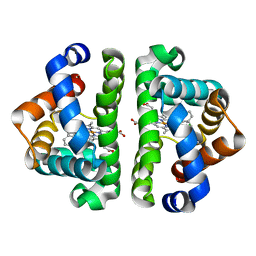 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloromethyl benzene bound to the XE4 cavity | | Descriptor: | (chloromethyl)benzene, CARBON MONOXIDE, GLOBIN-1, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
7AES
 
 | | Human carbonic anhydrase II in complex with 2,3,5,6-tetrafluoro-N-methyl-4-propylsulfanyl-benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrakis(fluoranyl)-~{N}-methyl-4-propylsulfanyl-benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BICINE, ... | | Authors: | Paketuryte, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of secondary sulfonamide binding to carbonic anhydrases.
Eur.Biophys.J., 50, 2021
|
|
1VJ3
 
 | | STRUCTURAL STUDIES ON BIO-ACTIVE COMPOUNDS. CRYSTAL STRUCTURE AND MOLECULAR MODELING STUDIES ON THE PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COFACTOR COMPLEX WITH TAB, A HIGHLY SELECTIVE ANTIFOLATE. | | Descriptor: | ACETIC ACID N-[2-CHLORO-5-[6-ETHYL-2,4-DIAMINO-PYRIMID-5-YL]-PHENYL]-[BENZYL-TRIAZEN-3-YL]ETHYL ESTER, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Rak, D, Luft, J.R, Queener, S.F, Laughton, C.A, Malcolm, F.G. | | Deposit date: | 2004-01-13 | | Release date: | 2004-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies on bioactive compounds. 30. Crystal structure and molecular modeling studies on the Pneumocystis carinii dihydrofolate reductase cofactor complex with TAB, a highly selective antifolate.
Biochemistry, 39, 2000
|
|
1VYZ
 
 | | Structure of CDK2 complexed with PNU-181227 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(5-CYCLOPROPYL-1H-PYRAZOL-3-YL)BENZAMIDE | | Authors: | Pevarello, P, Brasca, M.G, Amici, R, Orsini, P, Traquandi, G, Corti, L, Piutti, C, Sansonna, P, Villa, M, Pierce, B.S, Pulici, M, Giordano, G, Martina, K, Lfritzen, E, Nugent, R.A, Casale, E, Cameron, A, Ciomei, M, Roletto, F, Isacchi, A, Fogliatto, G, Pesenti, E, Pastori, W, Marsiglio, W, Leach, K.L, Clare, P.M, Fiorentini, F, Varasi, M, Vulpetti, A, Warpehoski, M.A. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2/Cyclin a as Antitumor Agents. 1. Lead Finding
J.Med.Chem., 47, 2004
|
|
3G4W
 
 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chlorobenzene bound to the XE4 cavity | | Descriptor: | CARBON MONOXIDE, GLOBIN-1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
2RAZ
 
 | |
