2PYB
 
 | |
2XTV
 
 | |
2O2W
 
 | |
2O31
 
 | |
1H8Y
 
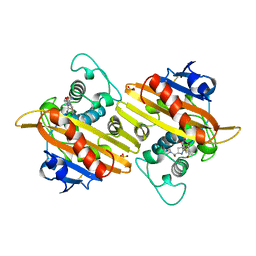 | | Crystal structure of the class D beta-lactamase OXA-13 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, BETA-LACTAMASE, SULFATE ION | | Authors: | Pernot, L, Frenois, F, Rybkine, T, L'Hermite, G, Petrella, S, Delettre, J, Jarlier, V, Collatz, E, Sougakoff, W. | | Deposit date: | 2001-02-17 | | Release date: | 2001-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Class D B-Lactamase Oxa-13 in the Native Form and in Complex with Meropenem
J.Mol.Biol., 310, 2001
|
|
2MSO
 
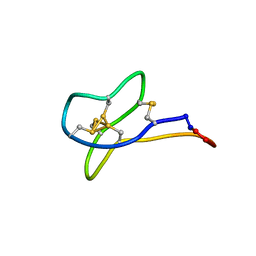 | | Solution study of cGm9a | | Descriptor: | Conotoxin Gm9.1 | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
2MSQ
 
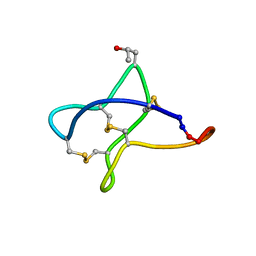 | | Solution study of cBru9a | | Descriptor: | Conotoxin cBru9a | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
1TYR
 
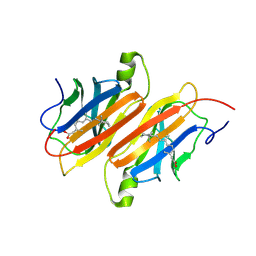 | | TRANSTHYRETIN COMPLEX WITH RETINOIC ACID | | Descriptor: | (9cis)-retinoic acid, TRANSTHYRETIN | | Authors: | Zanotti, G, D'Acunto, M.R, Malpeli, G, Folli, C, Berni, R. | | Deposit date: | 1995-05-12 | | Release date: | 1995-09-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the transthyretin--retinoic-acid complex
Eur.J.Biochem., 234, 1995
|
|
2J15
 
 | | Cyclic MrIA: An exceptionally stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter. | | Descriptor: | MAI126P | | Authors: | Lovelace, E.S, Armishaw, C.J, Colgrave, M.L, Walstrom, M.E, Alewood, P.F, Daly, N.L, Craik, D.J. | | Deposit date: | 2006-08-09 | | Release date: | 2006-11-01 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Cyclic MrIA: a stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter.
J. Med. Chem., 49, 2006
|
|
1O5H
 
 | |
2KCP
 
 | | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S8E; FROM Methanothermobacter thermautotrophicus, NORTHEASTSTRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET Tr71d | | Descriptor: | 30S ribosomal protein S8e | | Authors: | Liu, G, Rossi, P, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-24 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S8E FROM Methanothermobacter thermautotrophicus, NORTHEASTSTRUCTURAL GENOMICS CONSORTIUM (NESG) TARGET
To be Published
|
|
3EZ1
 
 | |
3I4J
 
 | | Crystal structure of Aminotransferase, class III from Deinococcus radiodurans | | Descriptor: | Aminotransferase, class III, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Aminotransferase, class III from Deinococcus radiodurans
To be Published
|
|
3I5T
 
 | | CRYSTAL STRUCTURE OF AMINOTRANSFERASE PRK07036 FROM Rhodobacter sphaeroides KD131 | | Descriptor: | Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AMINOTRANSFERASE PRK07036 FROM Rhodobacter sphaeroides
To be Published
|
|
3F0H
 
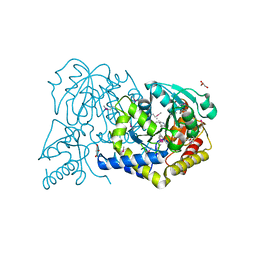 | |
2YRR
 
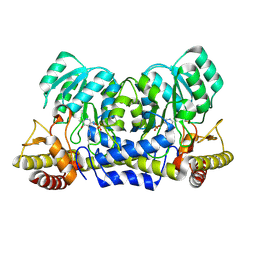 | | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8 | | Descriptor: | Aminotransferase, class V, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8
To be Published
|
|
3CXJ
 
 | |
4BEX
 
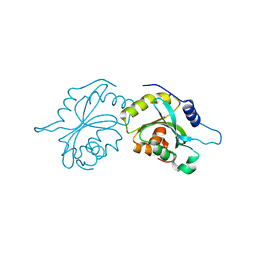 | | Structure of human Cofilin1 | | Descriptor: | COFILIN-1 | | Authors: | Klejnot, M, Gabrielsen, M, Cameron, J, Mleczak, A, Talapatra, S.K, Kozielski, F, Pannifer, A, Olson, M.F. | | Deposit date: | 2013-03-12 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the Human Cofilin1 Structure Reveals Conformational Changes Required for Actin-Binding
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6OTB
 
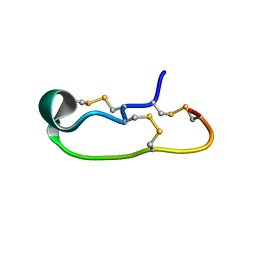 | |
2RAQ
 
 | | Crystal structure of the MTH889 protein from Methanothermobacter thermautotrophicus. Northeast Structural Genomics Consortium target TT205 | | Descriptor: | CALCIUM ION, Conserved protein MTH889 | | Authors: | Forouhar, F, Su, M, Xu, X, Seetharaman, J, Mao, L, Xiao, R, Ma, L.-C, Conover, K, Baran, M.C, Acton, T.B, Montelione, G.T, Arrowsmith, C.H, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-17 | | Release date: | 2007-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of the MTH889 protein from Methanothermobacter thermautotrophicus.
To be Published
|
|
6OTA
 
 | |
2M62
 
 | |
6CEI
 
 | |
1X0M
 
 | | a Human Kynurenine Aminotransferase II Homologue from Pyrococcus horikoshii OT3 | | Descriptor: | Aminotransferase II Homologue | | Authors: | Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2005-03-24 | | Release date: | 2005-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a human kynurenine aminotransferase II homologue from Pyrococcus horikoshii OT3 at 2.20 A resolution
Proteins, 61, 2005
|
|
5AIZ
 
 | | The PIAS-like coactivator Zmiz1 is a direct and selective cofactor of Notch1 in T-cell development and leukemia | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Cho, H.J, Murai, M, Chiang, M, Cierpicki, T. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-28 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Pias-Like Coactivator Zmiz1 is a Direct and Selective Cofactor of Notch1 in T-Cell Development and Leukemia
Immunity, 43, 2015
|
|
