7F1L
 
 | | Designed enzyme RA61 M48K/I72D mutant: form V | | Descriptor: | CHLORIDE ION, Engineered Retroaldolase, IMIDAZOLE | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1H
 
 | | Designed enzyme RA61 M48K/I72D mutant: form I | | Descriptor: | Engineered Retroaldolase, FORMIC ACID, GLYCEROL | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1J
 
 | | Designed enzyme RA61 M48K/I72D mutant: form III | | Descriptor: | Engineered Retroaldolase | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
5E28
 
 | | Crystal structure of human carbonic anhydrase II in complex with the 4-(4-aminophenyl)benzenesulfonamide inhibitor | | Descriptor: | 4'-aminobiphenyl-4-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 4-Arylbenzenesulfonamides as Human Carbonic Anhydrase Inhibitors (hCAIs): Synthesis by Pd Nanocatalyst-Mediated Suzuki-Miyaura Reaction, Enzyme Inhibition, and X-ray Crystallographic Studies.
J.Med.Chem., 59, 2016
|
|
1LLU
 
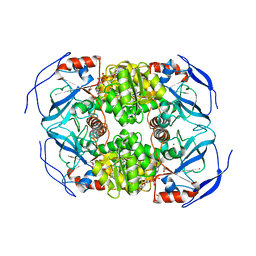 | | THE TERNARY COMPLEX OF PSEUDOMONAS AERUGINOSA ALCOHOL DEHYDROGENASE WITH ITS COENZYME AND WEAK SUBSTRATE | | Descriptor: | 1,2-ETHANEDIOL, Alcohol Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Levin, I, Meiri, G, Peretz, M, Frolow, F, Burstein, Y. | | Deposit date: | 2002-04-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The ternary complex of Pseudomonas aeruginosa alcohol dehydrogenase with NADH and ethylene glycol.
Protein Sci., 13, 2004
|
|
8QLP
 
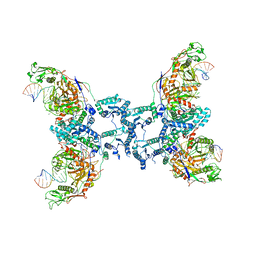 | | CryoEM structure of the RNA/DNA bound SPARTA (BabAgo/TIR-APAZ) tetrameric complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*C)-3'), MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
8QLO
 
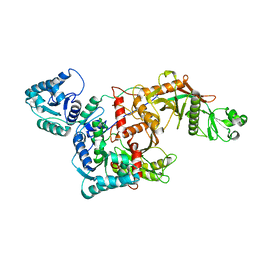 | | CryoEM structure of the apo SPARTA (BabAgo/TIR-APAZ) complex | | Descriptor: | Short prokaryotic Argonaute, Toll/interleukin-1 receptor domain-containing protein | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
5E2S
 
 | |
4AO6
 
 | | Native structure of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | ESTERASE | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
4AO7
 
 | | Zinc bound structure of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | ESTERASE, ZINC ION | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
5E2K
 
 | | Crystal structure of human carbonic anhydrase II in complex with the 4-(3-aminophenyl)benzenesulfonamide inhibitor | | Descriptor: | 3'-aminobiphenyl-4-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T. | | Deposit date: | 2015-10-01 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 4-Arylbenzenesulfonamides as Human Carbonic Anhydrase Inhibitors (hCAIs): Synthesis by Pd Nanocatalyst-Mediated Suzuki-Miyaura Reaction, Enzyme Inhibition, and X-ray Crystallographic Studies.
J.Med.Chem., 59, 2016
|
|
4AO8
 
 | | PEG-bound complex of a novel cold-adapted esterase from an Arctic intertidal metagenomic library | | Descriptor: | DI(HYDROXYETHYL)ETHER, ESTERASE | | Authors: | Fu, J, Leiros, H.-K.S, Pascale, D.d, Johnson, K.A, Blencke, H.M, Landfald, B. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and Structural Studies of a Novel Cold-Adapted Esterase from an Arctic Intertidal Metagenomic Library.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
5H2X
 
 | |
1XZ4
 
 | | Intersubunit Interactions Associated with Tyr42alpha Stabilize the Quaternary-T Tetramer but are not Major Quaternary Constraints in Deoxyhemoglobin: alphaY42A deoxyhemoglobin no-salt | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Rogers, P.H, Arnone, A, Hui, H.L, Wierzba, A, DeYoung, A, Kwiatkowski, L.D, Noble, R.W, Juszczak, L.J, Peterson, E.S, Friedman, J.M. | | Deposit date: | 2004-11-11 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intersubunit interactions associated with tyr42alpha stabilize the quaternary-T tetramer but are not major quaternary constraints in deoxyhemoglobin
Biochemistry, 44, 2005
|
|
5K4D
 
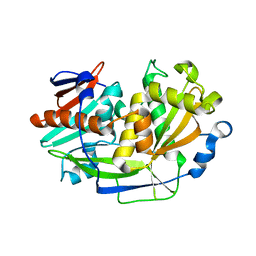 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 3 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
3UIQ
 
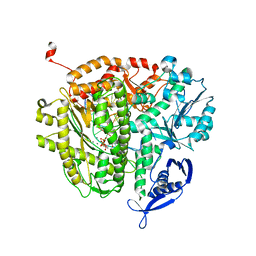 | | RB69 DNA Polymerase Ternary Complex containing dUpNpp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3', 5'-D(*TP*CP*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-11-05 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Bidentate and tridentate metal-ion coordination states within ternary complexes of RB69 DNA polymerase.
Protein Sci., 21, 2012
|
|
4BG4
 
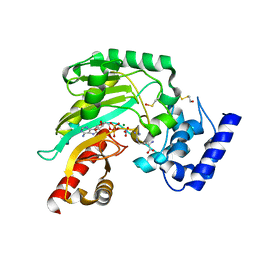 | | Crystal structure of Litopenaeus vannamei arginine kinase in a ternary analog complex with arginine, ADP-Mg and NO3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, ARGININE KINASE, ... | | Authors: | Lopez-Zavala, A.A, Garcia-Orozco, K.D, Carrasco-Miranda, J.S, Sugich-Miranda, R, Velazquez-Contreras, E.F, Criscitiello, M.F, GBrieba, L, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2013-03-22 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal Structure of Shrimp Arginine Kinase in Binary Complex with Arginine-A Molecular View of the Phosphagen Precursor Binding to the Enzyme.
J.Bioenerg.Biomembr., 45, 2013
|
|
2O3X
 
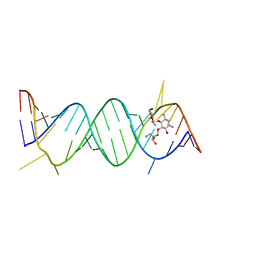 | | Crystal Structure of the Prokaryotic Ribosomal Decoding Site Complexed with Paromamine Derivative NB30 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-DIAMINO-2-[(5-AMINO-5-DEOXY-BETA-D-RIBOFURANOSYL)OXY]-3-HYDROXYCYCLOHEXYL 2-AMINO-2-DEOXY-ALPHA-D-GLUCOPYRANOSIDE, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
3OMW
 
 | | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II | | Descriptor: | CG14216 | | Authors: | Zhang, Y, Zhang, M, Zhang, Y. | | Deposit date: | 2010-08-27 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8701 Å) | | Cite: | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
2WL0
 
 | | HIV-1 Protease Inhibitors Containing a Tertiary Alcohol in the Transition-State Mimic with Improved Cell-Based Antiviral Activity | | Descriptor: | METHYL [(1S)-1-({2-[(3S)-3-BENZYL-3-HYDROXY-4-{[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]AMINO}-4-OXOBUTYL]-2-(4-PYRIDIN-2-YLBENZYL)HYDRAZINO}CARBONYL)-2,2-DIMETHYLPROPYL]CARBAMATE, PROTEASE | | Authors: | Mahalingam, A.K, Axelsson, L, Ekegren, J.K, Wannberg, J, Kihlstrom, J, Wallberg, H, Samuelsson, B, Larhed, M, Hallberg, A, Unge, T. | | Deposit date: | 2009-06-19 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HIV-1 Protease Inhibitors with a Transition-State Mimic Comprising a Tertiary Alcohol: Improved Antiviral Activity in Cells.
J.Med.Chem., 53, 2010
|
|
2WKZ
 
 | | HIV-1 Protease Inhibitors Containing a Tertiary Alcohol in the Transition-State Mimic with Improved Cell-Based Antiviral Activity | | Descriptor: | METHYL [(1S)-1-({2-[(3S)-3-BENZYL-3-HYDROXY-4-{[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]AMINO}-4-OXOBUTYL]-2-(4-PYRIDIN-2-YLBENZYL)HYDRAZINO}CARBONYL)-2,2-DIMETHYLPROPYL]CARBAMATE, PROTEASE | | Authors: | Mahalingam, A.K, Axelsson, L, Ekegren, J.K, Wannberg, J, Kihlstrom, J, Wallberg, H, Samuelsson, B, Larhed, M, Hallberg, A, Unge, T. | | Deposit date: | 2009-06-19 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | HIV-1 Protease Inhibitors with a Transition-State Mimic Comprising a Tertiary Alcohol: Improved Antiviral Activity in Cells.
J.Med.Chem., 53, 2010
|
|
5CTS
 
 | |
4RBN
 
 | | The crystal structure of Nitrosomonas europaea sucrose synthase: Insights into the evolutionary origin of sucrose metabolism in prokaryotes | | Descriptor: | Sucrose synthase:Glycosyl transferases group 1 | | Authors: | Wu, R, Asencion Diez, M.D, Figueroa, C.M, Machtey, M, Iglesias, A.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The Crystal Structure of Nitrosomonas europaea Sucrose Synthase Reveals Critical Conformational Changes and Insights into Sucrose Metabolism in Prokaryotes.
J.Bacteriol., 197, 2015
|
|
3NM1
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | 2-TRIFLUOROMETHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
3NL5
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
