4V41
 
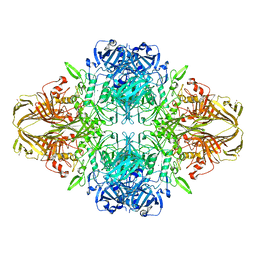 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
8HLA
 
 | |
2RSG
 
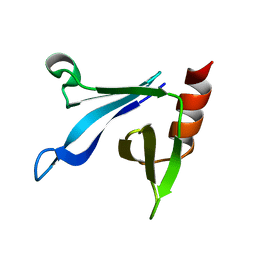 | | Solution structure of the CERT PH domain | | Descriptor: | Collagen type IV alpha-3-binding protein | | Authors: | Sugiki, T, Takeuchi, K, Tokunaga, Y, Kumagai, K, Kawano, M, Nishijima, M, Hanada, K, Takahashi, H, Shimada, I. | | Deposit date: | 2012-02-25 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the Golgi association by the pleckstrin homology domain of the ceramide trafficking protein (CERT)
J.Biol.Chem., 287, 2012
|
|
4V8A
 
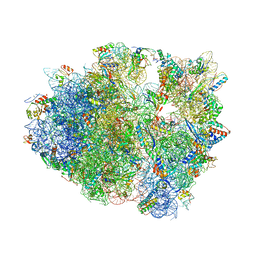 | | The structure of thermorubin in complex with the 70S ribosome from Thermus thermophilus. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D, Johnson, F.A, Steitz, T.A. | | Deposit date: | 2011-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibiotic thermorubin inhibits protein synthesis by binding to inter-subunit bridge b2a of the ribosome.
J.Mol.Biol., 416, 2012
|
|
8HIX
 
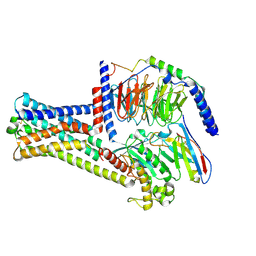 | | Cryo-EM structure of GPR21_m5_Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X, Xu, F. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
4V4R
 
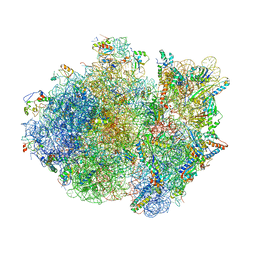 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-09-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
2RU8
 
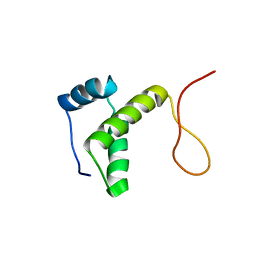 | | DnaT C-terminal domain | | Descriptor: | Primosomal protein 1 | | Authors: | Abe, Y, Tani, J, Fujiyama, S, Urabe, M, Sato, K, Aramaki, T, Katayama, T, Ueda, T. | | Deposit date: | 2014-01-29 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of the primosome protein DnaT-functional structures for homotrimerization, dissociation of ssDNA from the PriB·ssDNA complex, and formation of the DnaT·ssDNA complex.
Febs J., 281, 2014
|
|
8H3E
 
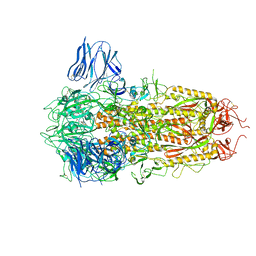 | | Complex structure of a small molecule (SPC-14) bound SARS-CoV-2 spike protein, closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
8H7H
 
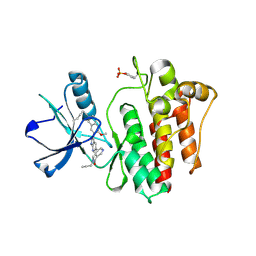 | | The crystal structure of human abl1 kinase domain in complex with abl1-A-EBA | | Descriptor: | 5-[3-(6-methoxyisoquinolin-7-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N-methyl-N-prop-2-ynyl-pyridine-3-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C.J, Zhang, Z.M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27789712 Å) | | Cite: | 2-Ethynylbenzaldehyde-Based, Lysine-Targeting Irreversible Covalent Inhibitors for Protein Kinases and Nonkinases.
J.Am.Chem.Soc., 2023
|
|
2RRQ
 
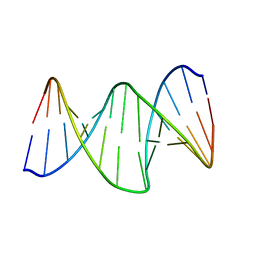 | | DNA oligomer containing propylene cross-linked cyclic 2'-deoxyuridylate dimer | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*CP*AP*(JDT)P*TP*AP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*TP*AP*AP*TP*GP*AP*AP*GP*G)-3') | | Authors: | Furuita, K, Murata, S, Jee, J.G, Ichikawa, S, Matsuda, A, Kojima, C. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Feature of Bent DNA Recognized by HMGB1
J.Am.Chem.Soc., 133, 2011
|
|
5SPU
 
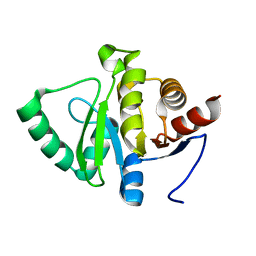 | |
4V8R
 
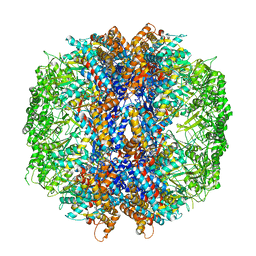 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Kalisman, N, Schroder, G.F, Levitt, M. | | Deposit date: | 2012-03-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
5SPT
 
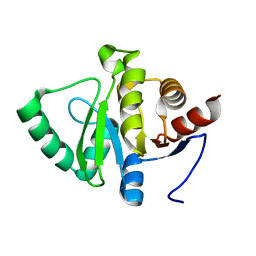 | |
4V8Y
 
 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
8HB8
 
 | |
2RIP
 
 | | Structure of DPPIV in complex with an inhibitor | | Descriptor: | (3R,4R)-4-(pyrrolidin-1-ylcarbonyl)-1-(quinoxalin-2-ylcarbonyl)pyrrolidin-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Qiu, X. | | Deposit date: | 2007-10-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design and synthesis of potent amido- and benzyl-substituted cis-3-amino-4-(2-cyanopyrrolidide)pyrrolidinyl DPP-IV inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8HH0
 
 | |
5SPS
 
 | |
4V9K
 
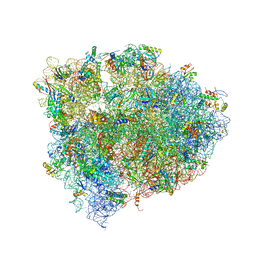 | | 70S ribosome translocation intermediate GDPNP-I containing elongation factor EFG/GDPNP, mRNA, and tRNA bound in the pe*/E state. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhou, J, Lancaster, L, Donohue, J.P, Noller, H.F. | | Deposit date: | 2013-04-24 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation.
Science, 340, 2013
|
|
8GYX
 
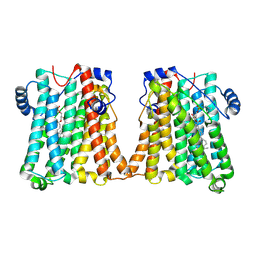 | | Cryo-EM structure of human CEPT1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Choline/ethanolaminephosphotransferase 1, MAGNESIUM ION | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for catalysis of human choline/ethanolamine phosphotransferase 1.
Nat Commun, 14, 2023
|
|
4V0V
 
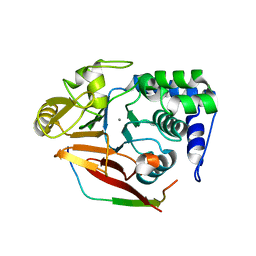 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-660) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT, ... | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
8C38
 
 | |
2RT6
 
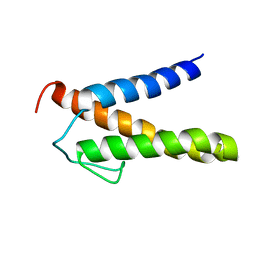 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for PriC N-terminal domain | | Descriptor: | Primosomal replication protein N'' | | Authors: | Aramaki, T, Abe, Y, Katayama, T, Ueda, T. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a replication restart primosome factor, PriC, in Escherichia coli.
Protein Sci., 22, 2013
|
|
4UVC
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Phenyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4S)-5-[5-[(1S)-1-azanyl-1,3-diphenyl-propyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-4aH-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
8HCX
 
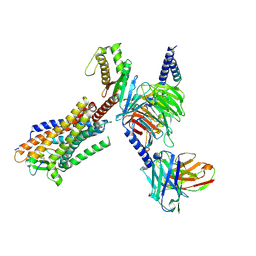 | | Cryo-EM structure of Endothelin1-bound ETBR-Gq complex | | Descriptor: | Endothelin receptor type B,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, Q, Jiang, Y, Xu, H.E, Ji, Y, Duan, J. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of peptide recognition and activation of endothelin receptors.
Nat Commun, 14, 2023
|
|
