2B1C
 
 | |
1GKU
 
 | |
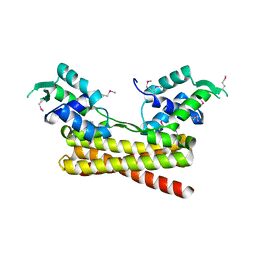
6JYI
 
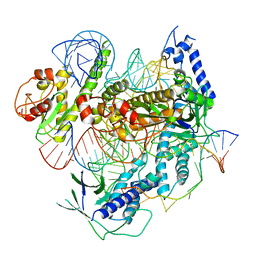
 | | Crystal structure of the PadR-like transcriptional regulator BC1756 from Bacillus cereus | | Descriptor: | Transcriptional repressor PadR | | Authors: | Kim, T.H, Park, S.C, Lee, K.C, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and DNA-binding studies of the PadR-like transcriptional regulator BC1756 from Bacillus cereus.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
7C7L
 
 | | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | | Descriptor: | CRISPR-associated protein Cas14a.1, DNA (40-mer), ZINC ION, ... | | Authors: | Takeda, N.S, Nakagawa, R, Okazaki, S, Hirano, H, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the miniature type V-F CRISPR-Cas effector enzyme.
Mol.Cell, 81, 2021
|
|
6TUP
 
 | |
7XHO
 
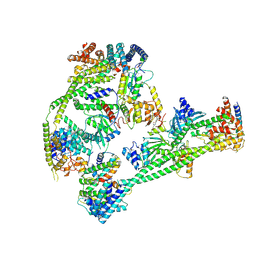 | | Structure of human inner kinetochore CCAN complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Tian, T, Wang, C.L, Yang, Z.S, Sun, L.F, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
2VKC
 
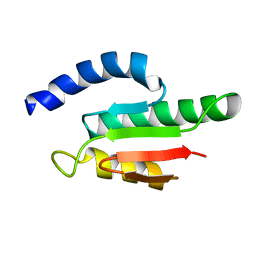 | | Solution structure of the B3BP Smr domain | | Descriptor: | NEDD4-BINDING PROTEIN 2 | | Authors: | Diercks, T, Ab, E, Daniels, M.A, deJong, R.N, Besseling, R, Kaptein, R, Folkers, G.E. | | Deposit date: | 2007-12-18 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the DNA- Binding Activity of the B3BP-Smr Domain.
J.Mol.Biol., 383, 2008
|
|
4CEJ
 
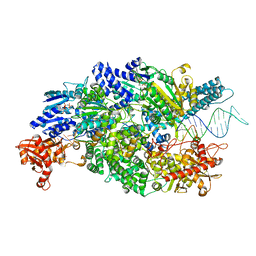 | | Crystal structure of AddAB-DNA-ADPNP complex at 3 Angstrom resolution | | Descriptor: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | Authors: | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | Deposit date: | 2013-11-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
7KPO
 
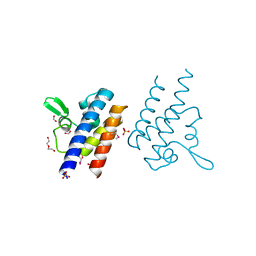 | | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Response regulator, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High Resolution Crystal Structure of the DNA-binding Domain from the Sensor Histidine Kinase ChiS from Vibrio cholerae.
To Be Published
|
|
6DPI
 
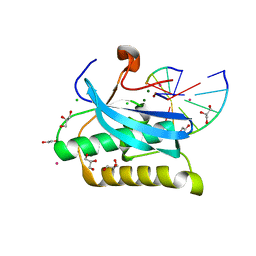 | |
4N6Q
 
 | | Crystal structure of VosA velvet domain | | Descriptor: | IODIDE ION, NITRATE ION, VosA | | Authors: | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
1KQ8
 
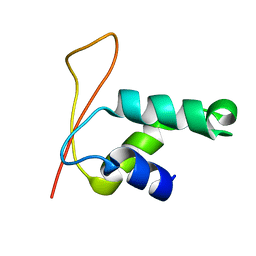 | | Solution Structure of Winged Helix Protein HFH-1 | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 3 FORKHEAD HOMOLOG 1 | | Authors: | Sheng, W, Rance, M, Liao, X. | | Deposit date: | 2002-01-04 | | Release date: | 2002-01-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure comparison of two conserved HNF-3/fkh proteins HFH-1 and genesis indicates the existence of folding differences in their complexes with a DNA binding sequence.
Biochemistry, 41, 2002
|
|
2KFS
 
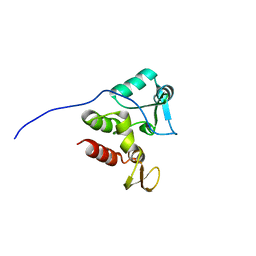 | | NMR structure of Rv2175c | | Descriptor: | Conserved hypothetical regulatory protein | | Authors: | Barthe, P, Cohen-Gonsaud, M, Roumestand, C, Molle, V. | | Deposit date: | 2009-02-27 | | Release date: | 2009-05-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The Mycobacterium tuberculosis Ser/Thr Kinase Substrate Rv2175c Is a DNA-binding Protein Regulated by Phosphorylation.
J.Biol.Chem., 284, 2009
|
|
2L31
 
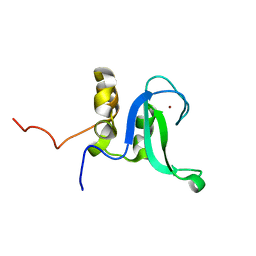 | | Human PARP-1 zinc finger 2 | | Descriptor: | Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Videler, H. | | Deposit date: | 2010-08-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger.
J.Mol.Biol., 407, 2011
|
|
2L30
 
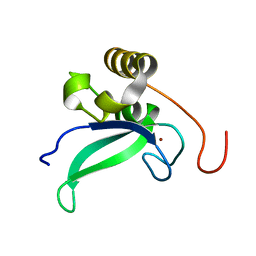 | | Human PARP-1 zinc finger 1 | | Descriptor: | Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Videler, H. | | Deposit date: | 2010-08-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger.
J.Mol.Biol., 407, 2011
|
|
1EQZ
 
 | | X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION | | Descriptor: | 146 NUCLEOTIDES LONG DNA, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Hanson, B.L, Harp, J.M, Timm, D.E, Bunick, G.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Asymmetries in the nucleosome core particle at 2.5 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
8F27
 
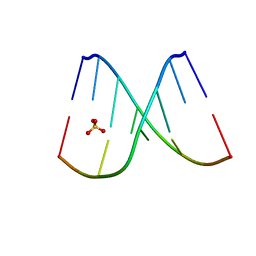 | | Mirror-image DNA containing 2'-OMe-L-uridine residue | | Descriptor: | Mirror-image DNA (5'-D(*(0DG)P*(0MU)P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DA)P*(0DC))-3'), SULFATE ION | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2022-11-07 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Derivatization of Mirror-Image l-Nucleic Acids with 2'-OMe Modification for Thermal and Structural Stabilization.
Chembiochem, 24, 2023
|
|
2LYH
 
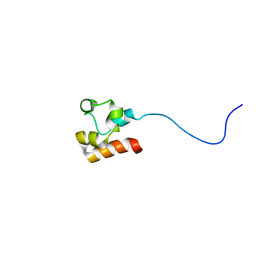 | | Structure of Faap24 residues 141-215 | | Descriptor: | Fanconi anemia-associated protein of 24 kDa | | Authors: | Wienk, H, Slootweg, J, Kaptein, R, Boelens, R, Folkers, G.E. | | Deposit date: | 2012-09-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Fanconi anemia associated protein FAAP24 uses two substrate specific binding surfaces for DNA recognition.
Nucleic Acids Res., 41, 2013
|
|
4L5R
 
 | |
8HWF
 
 | | Cryo-EM Structure of D5 ADP-ssDNA form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Li, Y.N, Zhu, J, Guo, Y.Y, Yan, R.H. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1DRG
 
 | | CRYSTAL STRUCTURE OF TRIMERIC CRE RECOMBINASE-LOX COMPLEX | | Descriptor: | 5'-D(*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3', 5'-D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*GP*C)-3', CRE RECOMBINASE | | Authors: | Woods, K.C, Baldwin, E.P. | | Deposit date: | 2000-01-06 | | Release date: | 2001-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction
J.Mol.Biol., 313, 2001
|
|
4MKP
 
 | | Crystal structure of human cGAS apo form | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural and Functional Analyses of DNA-Sensing and Immune Activation by Human cGAS
Plos One, 8, 2013
|
|
8A5A
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of INO80 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-like protein ARP8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8JMK
 
 | | Structure of Helicobacter pylori Soj mutant, D41A bound to DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
8A5O
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of S. cerevisiae INO80 | | Descriptor: | Actin, Actin-like protein ARP8, Actin-related protein 4, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
