1A1I
 
 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCAC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), RADR ZIF268 ZINC FINGER PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1G
 
 | | DSNR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GCGT SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), DSNR ZINC FINGER PEPTIDE, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1L
 
 | | ZIF268 ZINC FINGER-DNA COMPLEX (GCAC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
1A1K
 
 | | RADR (ZIF268 VARIANT) ZINC FINGER-DNA COMPLEX (GACC SITE) | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*CP*CP*AP*CP*GP*C)-3'), RADR ZIF268 VARIANT, ... | | Authors: | Elrod-Erickson, M, Benson, T.E, Pabo, C.O. | | Deposit date: | 1997-12-10 | | Release date: | 1998-06-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition.
Structure, 6, 1998
|
|
9LNQ
 
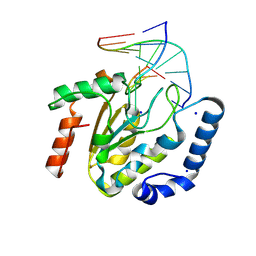 | | The hUNG bound to DNA product embedding 4primer-OCH3-dU | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(KBC)P*AP*TP*CP*TP*T)-3'), SODIUM ION, ... | | Authors: | Liu, Y, Zhou, C, Zhan, X, Fan, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
J.Am.Chem.Soc., 147, 2025
|
|
4X6A
 
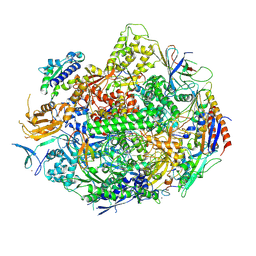 | | Crystal structure of yeast RNA polymerase II encountering oxidative Cyclopurine DNA lesions | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2014-12-07 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X67
 
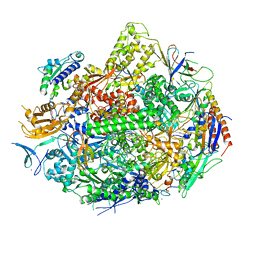 | | Crystal structure of elongating yeast RNA polymerase II stalled at oxidative Cyclopurine DNA lesions. | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2014-12-07 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7ICF
 
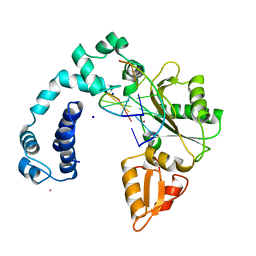 | |
7ICU
 
 | |
7ICJ
 
 | |
5NSR
 
 | | Cryo-EM structure of RNA polymerase-sigma54 holo enzyme with promoter DNA closed complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Glyde, R, Ye, F.Z, Darbari, V.C, Zhang, N, Buck, M, Zhang, X.D. | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of RNA Polymerase Closed and Intermediate Complexes Reveal Mechanisms of DNA Opening and Transcription Initiation.
Mol. Cell, 67, 2017
|
|
1BF4
 
 | | CHROMOSOMAL DNA-BINDING PROTEIN SSO7D/D(GCGAACGC) COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*5IUP*CP*GP*C)-3'), PROTEIN (CHROMOSOMAL PROTEIN SSO7D) | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Padmanabhan, S, Lim, L, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1998-05-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA.
Nat.Struct.Biol., 5, 1998
|
|
9NEA
 
 | | Human polymerase epsilon bound to PCNA and DNA with a pre-existing mismatch in the blocked conformation II | | Descriptor: | DNA (29-MER), DNA (35-MER), DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | The proofreading mechanism of the human leading-strand DNA polymerase epsilon holoenzyme.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9NE9
 
 | | Human polymerase epsilon bound to PCNA and DNA with a pre-existing mismatch in the blocked conformation I | | Descriptor: | DNA (35-MER), DNA (49-MER), DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | The proofreading mechanism of the human leading-strand DNA polymerase epsilon holoenzyme.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
4CKK
 
 | | Apo structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit | | Descriptor: | DNA GYRASE SUBUNIT A | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
9NE7
 
 | | Human polymerase epsilon bound to PCNA and DNA with an in-situ-generated mismatch in the Pol-backtracking state | | Descriptor: | DNA (33-MER), DNA (47-MER), DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | The proofreading mechanism of the human leading-strand DNA polymerase epsilon holoenzyme.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9NE6
 
 | | Human polymerase epsilon bound to PCNA and DNA with an in-situ-generated mismatch in the mismatch-editing state | | Descriptor: | DNA (33-MER), DNA (47-MER), DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | The proofreading mechanism of the human leading-strand DNA polymerase epsilon holoenzyme.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9NE8
 
 | | Human polymerase epsilon bound to PCNA and DNA with an in-situ-generated mismatch in the mismatch-locking state | | Descriptor: | DNA (33-MER), DNA (47-MER), DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2025-02-19 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The proofreading mechanism of the human leading-strand DNA polymerase epsilon holoenzyme.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
5KE6
 
 | | mouse Klf4 ZnF1-3 and TpG/CpA sequence DNA complex structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*AP*CP*CP*TP*C)-3'), Krueppel-like factor 4, ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Distinctive Klf4 mutants determine preference for DNA methylation status.
Nucleic Acids Res., 44, 2016
|
|
5KEB
 
 | |
7SGL
 
 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
9MVY
 
 | | Crystal structure of ZMET2 in complex with unmethylated CTG DNA and a histone H3Kc9me2 peptide | | Descriptor: | C49 ssDNA, DNA (cytosine-5)-methyltransferase 1, Histone H3.2, ... | | Authors: | Herle, G, Fang, J, Song, J. | | Deposit date: | 2025-01-16 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of an Unfavorable de Novo DNA Methylation Complex of Plant Methyltransferase ZMET2.
J.Mol.Biol., 437, 2025
|
|
4CKL
 
 | | Structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit with simocyclinone D8 bound | | Descriptor: | DNA GYRASE SUBUNIT A, SIMOCYCLINONE D8 | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
