4JRQ
 
 | |
6LTV
 
 | | Crystal Structure of I122A/I330A variant of S-adenosylmethionine synthetase from Cryptosporidium hominis in complex with ONB-SAM (2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | MAGNESIUM ION, S-adenosylmethionine synthase, TRIPHOSPHATE, ... | | Authors: | Singh, R.K, Michailidou, F, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Engineered SAM Synthetases for Enzymatic Generation of AdoMet Analogs with Photocaging Groups and Reversible DNA Modification in Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1IXY
 
 | | Ternary complex of T4 phage BGT with UDP and a 13 mer DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*TP*C)-3', 5'-D(*GP*AP*TP*AP*CP*TP*3DRP*AP*GP*AP*TP*AP*G)-3', ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2002-07-09 | | Release date: | 2002-12-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Base-flipping Mechanism for the T4 Phage beta-Glucosyltransferase and Identification of a
Transition-state Analog
J.Mol.Biol., 324, 2002
|
|
6LTW
 
 | | Crystal structure of Apo form of I122A/I330A variant of S-adenosylmethionine synthetase from Cryptosporidium hominis | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, S-adenosylmethionine synthase | | Authors: | Singh, R.K, Michailidou, F, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineered SAM Synthetases for Enzymatic Generation of AdoMet Analogs with Photocaging Groups and Reversible DNA Modification in Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7Z9G
 
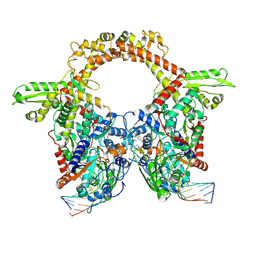 | | E.coli gyrase holocomplex with 217 bp DNA and Albi-2 | | Descriptor: | 4-[[3-(2-azanylethoxy)-2-oxidanyl-4-[[5-[[(2~{S})-2-[[4-[(6-oxidanylnaphthalen-2-yl)carbonylamino]phenyl]carbonylamino]-3-(1~{H}-1,2,3-triazol-4-yl)propanoyl]amino]pyridin-2-yl]carbonylamino]phenyl]carbonylamino]-3-methoxy-2-oxidanyl-benzoic acid, DNA (5'-D(*AP*AP*TP*CP*AP*CP*CP*CP*GP*CP*AP*CP*AP*GP*AP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*TP*TP*AP*TP*GP*CP*CP*TP*GP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Ghilarov, D, Heddle, J.G.H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin.
Nat Catal, 6, 2023
|
|
7Z9M
 
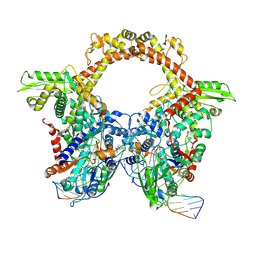 | | E.coli gyrase holocomplex with 217 bp DNA and Albi-1 (site AA) | | Descriptor: | 4-[[4-[[5-[[(2S)-2-[[5-[(4-cyanophenyl)carbonylamino]pyridin-2-yl]carbonylamino]-3-(1H-1,2,3-triazol-4-yl)propanoyl]amino]pyridin-2-yl]carbonylamino]-2-oxidanyl-3-propan-2-yloxy-phenyl]carbonylamino]benzoic acid, DNA (5'-D(*AP*AP*TP*CP*AP*CP*CP*CP*GP*CP*AP*CP*AP*GP*AP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*TP*TP*AP*TP*GP*CP*CP*TP*GP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Ghilarov, D, Heddle, J.G.H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin.
Nat Catal, 6, 2023
|
|
7Z9K
 
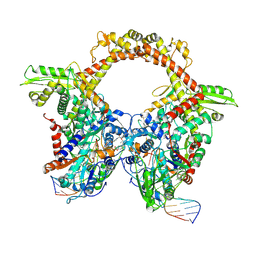 | | E.coli gyrase holocomplex with 217 bp DNA and Albi-1 (site TG) | | Descriptor: | 4-[[4-[[5-[[(2S)-2-[[5-[(4-cyanophenyl)carbonylamino]pyridin-2-yl]carbonylamino]-3-(1H-1,2,3-triazol-4-yl)propanoyl]amino]pyridin-2-yl]carbonylamino]-2-oxidanyl-3-propan-2-yloxy-phenyl]carbonylamino]benzoic acid, DNA (5'-D(*AP*AP*TP*CP*AP*CP*CP*CP*GP*CP*AP*CP*AP*GP*AP*TP*TP*T)-3'), DNA (5'-D(*GP*AP*TP*TP*TP*TP*AP*TP*GP*CP*CP*TP*GP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Ghilarov, D, Heddle, J.G.H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular mechanism of topoisomerase poisoning by the peptide antibiotic albicidin.
Nat Catal, 6, 2023
|
|
7KZR
 
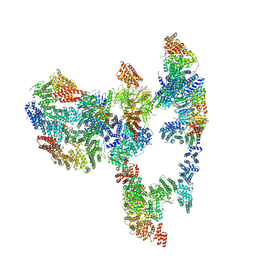 | | Structure of the human Fanconi Anaemia Core-UBE2T-ID complex | | Descriptor: | E3 ubiquitin-protein ligase FANCL, Fanconi anemia core complex-associated protein 100, Fanconi anemia core complex-associated protein 20, ... | | Authors: | Wang, S.L, Pavletich, N.P. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the FA core ubiquitin ligase closing the ID clamp on DNA.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KZQ
 
 | | Structure of the human Fanconi anaemia Core-ID complex | | Descriptor: | E3 ubiquitin-protein ligase FANCL, Fanconi anemia core complex-associated protein 100, Fanconi anemia core complex-associated protein 20, ... | | Authors: | Wang, S.L, Pavletich, N.P. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the FA core ubiquitin ligase closing the ID clamp on DNA.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5U34
 
 | | Crystal structure of AacC2c1-sgRNA binary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, sgRNA | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.255 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
2MXE
 
 | |
8Q3W
 
 | | ATP-bound IstB in complex to duplex DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (48-MER) Target DNA FW, DNA (48-MER) Traget DNA Rv, ... | | Authors: | de la Gandara, A, Spinola-Amilibia, M, Araujo-Bazan, L, Nunez-Ramirez, R, Berger, J.M, Arias-Palomo, E. | | Deposit date: | 2023-08-04 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular basis for transposase activation by a dedicated AAA+ ATPase.
Nature, 630, 2024
|
|
6W10
 
 | |
1NNE
 
 | | Crystal Structure of the MutS-ADPBeF3-DNA complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*GP*CP*GP*AP*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*TP*CP*GP*TP*C)-3', 5'-D(P*GP*GP*AP*CP*GP*AP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*GP*TP*CP*G)-3', ... | | Authors: | Alani, E, Lee, J.Y, Schofield, M.J, Kijas, A.W, Hsieh, P, Yang, W. | | Deposit date: | 2003-01-13 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure and biochemical analysis of the MutS-ADP-Beryllium Fluoride complex
suggests a conserved mechanism for ATP interactions in mismatch repair
J.Biol.Chem., 278, 2003
|
|
7BNR
 
 | |
7BNK
 
 | |
6WAA
 
 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and compound 34 (7-[(1S,5R)-1-amino-3-azabicyclo[3.1.0]hexan-3-yl]-4-(aminomethyl)-1-cyclopropyl-3,6-difluoro-8-methylquinolin-2(1H)-one) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(1S,5R)-1-amino-3-azabicyclo[3.1.0]hexan-3-yl]-4-(aminomethyl)-1-cyclopropyl-3,6-difluoro-8-methylquinolin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Topoisomerase Inhibitors Addressing Fluoroquinolone Resistance in Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
3MWY
 
 | |
5ZMM
 
 | | Structure of the Type IV phosphorothioation-dependent restriction endonuclease ScoMcrA | | Descriptor: | SULFATE ION, Uncharacterized protein McrA, ZINC ION | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
6WG4
 
 | |
6QTK
 
 | | 2.31A structure of gepotidacin with S.aureus DNA gyrase and doubly nicked DNA | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*G*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D. | | Deposit date: | 2019-02-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Mechanistic and Structural Basis for the Actions of the Antibacterial Gepotidacin against Staphylococcus aureus Gyrase.
Acs Infect Dis., 5, 2019
|
|
1CG7
 
 | | HMG PROTEIN NHP6A FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PROTEIN (NON HISTONE PROTEIN 6 A) | | Authors: | Allain, F.H.T, Yen, Y.M, Masse, J.E, Schultze, P, Dieckmann, T, Johnson, R.C, Feigon, J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding.
EMBO J., 18, 1999
|
|
7MG2
 
 | |
7MG3
 
 | |
7MG4
 
 | |
