8RHB
 
 | | Microtubule-associated kinesin-1 tail complex bound to AMPPNP, single-headed state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kif5b Tail, ... | | Authors: | Atherton, J, Chegkazi, M.S, Steiner, R.A. | | Deposit date: | 2023-12-15 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Microtubule association induces a Mg-free apo-like ADP pre-release conformation in kinesin-1 that is unaffected by its autoinhibitory tail.
Nat Commun, 16, 2025
|
|
9CV9
 
 | | Bufavirus 1 at pH 4.0 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-28 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
7B45
 
 | | Notum complex with ARUK3003934 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(3-methylsulfanylphenyl)sulfanyl-2~{H}-[1,2,4]triazolo[4,3-b]pyridazin-3-one, ... | | Authors: | Fish, P, Jones, E.Y. | | Deposit date: | 2020-12-02 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
8RHH
 
 | | Microtubule-associated kinesin-1 tail complex bound to AMPPNP, two-headed state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Atherton, J, Chegkazi, M.S, Steiner, R.A. | | Deposit date: | 2023-12-15 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Microtubule association induces a Mg-free apo-like ADP pre-release conformation in kinesin-1 that is unaffected by its autoinhibitory tail.
Nat Commun, 16, 2025
|
|
7B8N
 
 | | Notum-Fragment 197 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-azanyl-2-(pyridin-3-ylmethylamino)benzoic acid, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B4X
 
 | | Notum complex with ARUK3002697 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-methylphenyl)sulfanyl-2~{H}-[1,2,4]triazolo[4,3-b]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-02 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
6HCZ
 
 | |
6FAO
 
 | | Discovery and characterization of a thermostable GH6 endoglucanase from a compost metagenome | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 6, SULFATE ION | | Authors: | Jensen, M.S, Fredriksen, L, MacKenzie, A.K, Pope, P.B, Chylenski, P, Leiros, I, Williamson, A.K, Christopeit, T, Ostby, H, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and characterization of a thermostable two-domain GH6 endoglucanase from a compost metagenome.
PLoS ONE, 13, 2018
|
|
9CUZ
 
 | | Bufavirus 1 complexed with 6SLN | | Descriptor: | N-acetyl-alpha-neuraminic acid, VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-27 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
6ZIV
 
 | | Crystal structure of a Beta-glucosidase from Alicyclobacillus acidiphilus | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, ... | | Authors: | Gourlay, L.J. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Producing natural vanilla extract from green vanilla beans using a beta-glucosidase from Alicyclobacillus acidiphilus.
J.Biotechnol., 329, 2021
|
|
8K32
 
 | |
6SYI
 
 | |
5N1P
 
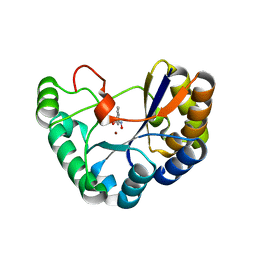 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with N-hydroxynaphthalene-1-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Peptidoglycan N-acetylglucosamine deacetylase, SODIUM ION, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
9FOP
 
 | | Half-closed CODH/ACS (Class 1) in the methylated state | | Descriptor: | CO-dehydrogenase, CO-methylating acetyl-CoA synthase, Fe(3)-Ni(1)-S(4) cluster, ... | | Authors: | Ruickoldt, J, Wendler, P, Dobbek, H. | | Deposit date: | 2024-06-12 | | Release date: | 2025-06-25 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Ligand binding to a Ni-Fe cluster orchestrates conformational changes of the CO-dehydrogenase-acetyl-CoA synthase complex.
Nat Catal, 8, 2025
|
|
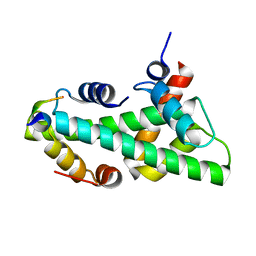
7BP6
 
 | |
6X3N
 
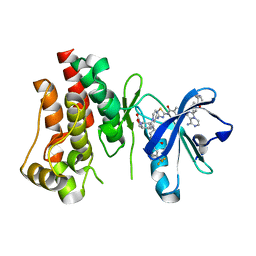 | | Co-structure of BTK kinase domain with L-005085737 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxohexahydro-3H-[1,3]oxazolo[3,4-a]pyridin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
9D6D
 
 | |
6THI
 
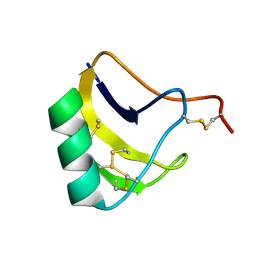 | | Solution structure of MeuNaTxalpha-1 toxin from Mesobuthus Eupeus | | Descriptor: | Sodium channel neurotoxin MeuNaTxalpha-1 | | Authors: | Mineev, K.S, Kuzmenkov, A.I, Khusainov, G.A, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of MeuNaTx alpha-1 toxin from scorpion venom highlights the importance of the nest motif.
Proteins, 2021
|
|
5NQF
 
 | | Crystal structure of Plasmodium falciparum AMA1 in complex with a 39 aa PvRON2 peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Vulliez-le Normand, B, Saul, F.A, Faber, B.W, Bentley, G.A. | | Deposit date: | 2017-04-20 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-reactivity between apical membrane antgen 1 and rhoptry neck protein 2 in P. vivax and P. falciparum: A structural and binding study.
PLoS ONE, 12, 2017
|
|
6H79
 
 | |
8AV0
 
 | | small molecule stabilizer (compound 1) for C-RAF pS259 and 14-3-3 | | Descriptor: | 1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-4-carboxamide, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
8EVJ
 
 | | CX3CR1 nucleosome bound PU.1 and C/EBPa | | Descriptor: | DNA (167-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Guan, R, Bai, Y, Lian, T. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBP alpha.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EVI
 
 | | CX3CR1 nucleosome and PU.1 complex containing disulfide bond mutations | | Descriptor: | DNA (167-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBP alpha.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5F8Z
 
 | | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1 | | Descriptor: | CYS-PRO-ALA-ARG-PHE-M70-ALA-LEU-PHE-CYS, Plasma kallikrein LIGHT CHAIN, SULFATE ION, ... | | Authors: | Xu, M, Jiang, L, Xu, P, Luo, Z, Andreasen, P, Huang, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1
To Be Published
|
|
7RXR
 
 | | Crystal Structure of BRD4(D1) with 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
