7OWZ
 
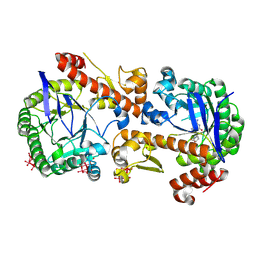 | | Heterodimeric murine tRNA-guanine transglycosylase in complex with queuine and in the presence of Anderson-Evans type (TEW) and Strandberg type polyoxometalate (POM) | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-tungstotellurate(VI), Queuine tRNA-ribosyltransferase accessory subunit 2, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
1ZU0
 
 | |
1F2W
 
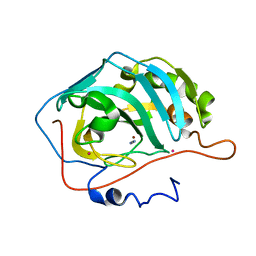 | | THE MECHANISM OF CYANAMIDE HYDRATION CATALYZED BY CARBONIC ANHYDRASE II REVEALED BY CRYOGENIC X-RAY DIFFRACTION | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, CARBONIC ANHYDRASE II, CYANAMIDE, ... | | Authors: | Guerri, A, Briganti, F, Scozzafava, A, Supuran, C.T, Mangani, S. | | Deposit date: | 2000-05-30 | | Release date: | 2000-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of cyanamide hydration catalyzed by carbonic anhydrase II suggested by cryogenic X-ray diffraction.
Biochemistry, 39, 2000
|
|
2Z7S
 
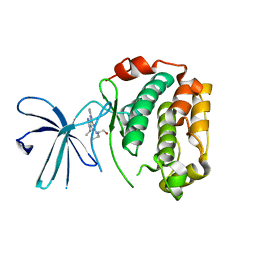 | | Crystal Structure of the N-terminal Kinase Domain of Human RSK1 bound to Purvalnol A | | Descriptor: | 2-({6-[(3-CHLOROPHENYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)-3-METHYLBUTAN-1-OL, Ribosomal protein S6 kinase alpha-1 | | Authors: | Ikuta, M, Munshi, S.K. | | Deposit date: | 2007-08-28 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the N-terminal kinase domain of human RSK1 bound to three different ligands: Implications for the design of RSK1 specific inhibitors.
Protein Sci., 16, 2007
|
|
6WGM
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for pyroglutamate (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, scf7180008839099) in complex with co-purified pyroglutamate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Fedorov, E, Vetting, M.W, Hogle, S.L, Dupont, C.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2020-04-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, GOS_140), in complex with co-purified pyroglutamate
To Be Published
|
|
8H6H
 
 | |
8GKO
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | 1,2-ETHANEDIOL, Alkaline protease 1, FORMYL GROUP, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
8GKQ
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | 1,2-ETHANEDIOL, Alkaline protease 1, CALCIUM ION, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
8GKP
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | Alkaline protease 1, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
6X2M
 
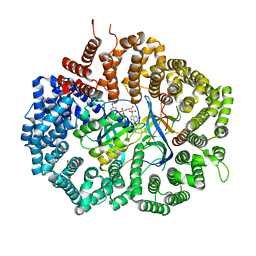 | | Crystal Structure of unliganded CRM1-Ran-RanBP1 | | Descriptor: | Exportin-1, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
5FOQ
 
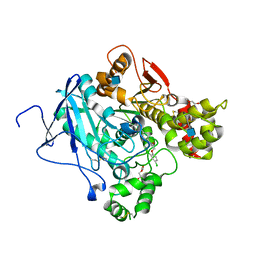 | | Acetylcholinesterase in complex with C7653 | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2,4-dichlorophenoxy)-N-[4-(1-piperidinylmethyl)phenyl]acetamide, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Berg, L, Mishra, B.K, Andersson, D.C, Ekstrom, F, Linusson, A. | | Deposit date: | 2015-11-25 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Nature of Activated Non-Classical Hydrogen Bonds: A Case Study on Acetylcholinesterase-Ligand Complexes.
Chemistry, 22, 2016
|
|
8VY3
 
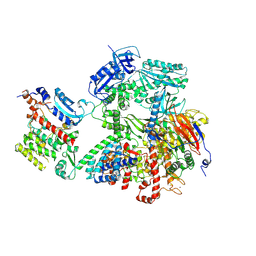 | | Human DNA polymerase alpha/primase - AavLEA1 (1:40 molar ratio) | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | Authors: | Abe, K.M, Li, G, Grant, T, Lim, C.J. | | Deposit date: | 2024-02-06 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Small LEA proteins mitigate air-water interface damage to fragile cryo-EM samples during plunge freezing.
Nat Commun, 15, 2024
|
|
5G1A
 
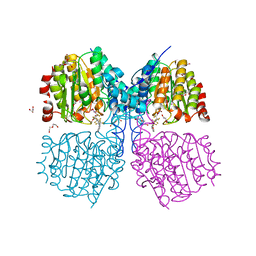 | | Bordetella Alcaligenes HDAH bound to PFSAHA | | Descriptor: | 2,2,3,3,4,4,5,5,6,6,7,7-dodecakis(fluoranyl)-~{N}-oxidanyl-~{N}'-phenyl-octanediamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
8T52
 
 | |
8T5A
 
 | |
6VJY
 
 | | Cryo-EM structure of Hrd1/Hrd3 monomer | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
8S9Q
 
 | |
2FEU
 
 | | P450CAM from Pseudomonas putida reconstituted with manganic protoporphyrin IX | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.G, Schlichting, I. | | Deposit date: | 2005-12-16 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The status of high-valent metal oxo complexes in the P450 cytochromes.
J.Inorg.Biochem., 100, 2006
|
|
8W39
 
 | |
8W8V
 
 | | High-resolution X-ray structure of cellulase Cel6A from Phanerochaete chrysosporium at cryogenic temperature, Enzyme-Product complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Glucanase, ... | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-09-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W4Y
 
 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature, low-D2O-solvent | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.4 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W4W
 
 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.36 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W8U
 
 | | High-resolution X-ray structure of cellulase Cel6A from Phanerochaete chrysosporium at cryogenic temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-09-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W4X
 
 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature, Enzyme-Product complex | | Descriptor: | Glucanase, SODIUM ION, beta-D-glucopyranose, ... | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.4 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W4Z
 
 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature, Enzyme-Product complex, H2O solvent | | Descriptor: | Glucanase, SODIUM ION, beta-D-glucopyranose, ... | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-30 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
