4H5S
 
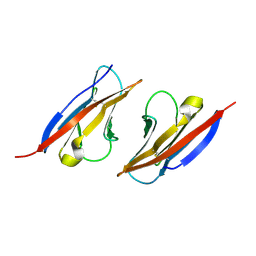 | | Complex structure of Necl-2 and CRTAM | | Descriptor: | Cell adhesion molecule 1, Cytotoxic and regulatory T-cell molecule | | Authors: | Zhang, S, Lu, G, Qi, J, Li, Y, Zhang, Z, Zhang, B, Yan, J, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-08-07 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Competition of cell adhesion and immune recognition: insights into the interaction between CRTAM and nectin-like 2.
Structure, 21, 2013
|
|
4HA5
 
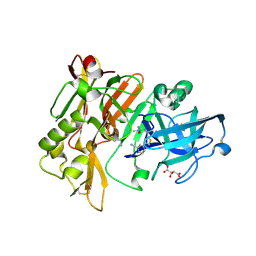 | | Structure of BACE Bound to (S)-3-(5-(2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl)thiophen-3-yl)benzonitrile | | Descriptor: | 3-{5-[(2E,4S)-2-imino-1,4-dimethyl-6-oxohexahydropyrimidin-4-yl]thiophen-3-yl}benzonitrile, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Mandal, M. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-17 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and Validation of Bicyclic Iminopyrimidinones As Beta Amyloid Cleaving Enzyme-1 (BACE1) Inhibitors: Conformational Constraint to Favor a Bioactive Conformation.
J.Med.Chem., 55, 2012
|
|
4H3W
 
 | |
4GLM
 
 | | Crystal structure of the SH3 Domain of DNMBP protein [Homo sapiens] | | Descriptor: | Dynamin-binding protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Guan, X, Huang, H, Tempel, W, Gu, J, Sidhu, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-14 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SH3 Domain of DNMBP protein [Homo sapiens]
to be published
|
|
4GT6
 
 | |
4GUA
 
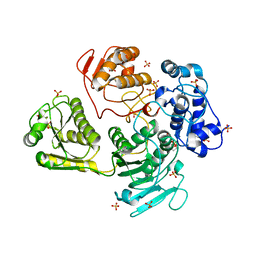 | | Alphavirus P23pro-zbd | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural polyprotein, SULFATE ION, ... | | Authors: | Shin, G, Yost, S, Miller, M, Marcotrigiano, J. | | Deposit date: | 2012-08-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structural and functional insights into alphavirus polyprotein processing and pathogenesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GVA
 
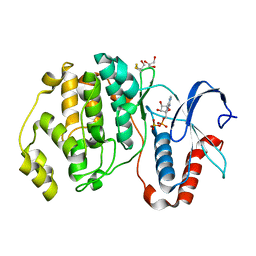 | |
4IVE
 
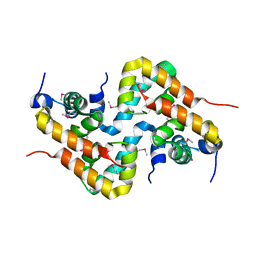 | |
4IWX
 
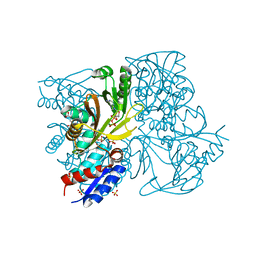 | | Rimk structure at 2.85A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Ribosomal protein S6 modification protein, ... | | Authors: | Shi, D, Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structure and function of Escherichia coli RimK, an ATP-grasp fold, l-glutamyl ligase enzyme.
Proteins, 81, 2013
|
|
4J1G
 
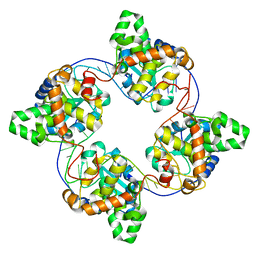 | | Leanyer orthobunyavirus nucleoprotein-ssRNA complex | | Descriptor: | Nucleocapsid, RNA (45-MER) | | Authors: | Niu, F, Shaw, N, Wang, Y, Jiao, L, Ding, W, Li, X, Zhu, P, Upur, H, Ouyang, S, Cheng, G, Liu, Z.J. | | Deposit date: | 2013-02-01 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structure of the Leanyer orthobunyavirus nucleoprotein-RNA complex reveals unique architecture for RNA encapsidation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IZ5
 
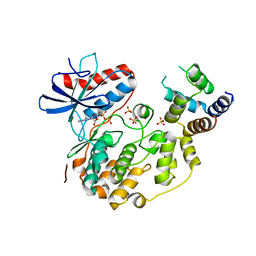 | | Structure of the complex between ERK2 phosphomimetic mutant and PEA-15 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Astrocytic phosphoprotein PEA-15, Mitogen-activated protein kinase 1, ... | | Authors: | Mace, P.D, Robinson, H, Riedl, S.J. | | Deposit date: | 2013-01-29 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of ERK2 bound to PEA-15 reveals a mechanism for rapid release of activated MAPK.
Nat Commun, 4, 2013
|
|
4J4Q
 
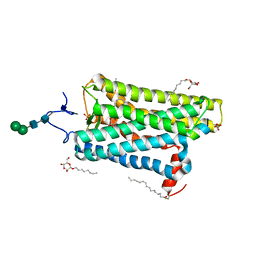 | | Crystal structure of active conformation of GPCR opsin stabilized by octylglucoside | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Park, J.H, Morizumi, T, Li, Y, Hong, J.E, Pai, E.F, Hofmann, K.P, Choe, H.W, Ernst, O.P. | | Deposit date: | 2013-02-07 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Opsin, a structural model for olfactory receptors?
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4IZ7
 
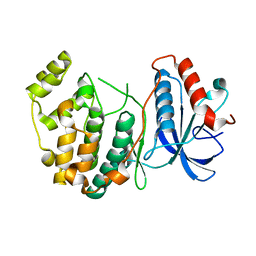 | |
4IYJ
 
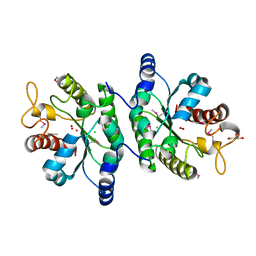 | |
4J3J
 
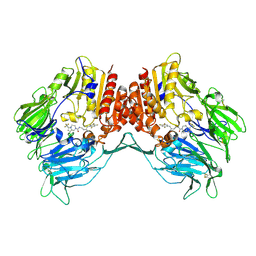 | | Crystal Structure of DPP-IV with Compound C3 | | Descriptor: | Dipeptidyl peptidase 4, N-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butyl]-6-(trifluoromethyl)-3,4-dihydropyrrolo[1,2-a]pyrazine-2(1H)-carboxamide | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 4-(2,4,5-trifluorophenyl)butane-1,3-diamines as dipeptidyl peptidase IV inhibitors
Chemmedchem, 8, 2013
|
|
4J7Q
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh3 complexed with phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein PDR16 | | Authors: | Yang, H, Im, Y.J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants for phosphatidylinositol recognition by Sfh3 and substrate-induced dimer-monomer transition during lipid transfer cycles.
Febs Lett., 587, 2013
|
|
4JD2
 
 | |
4HAG
 
 | | Crystal structure of fc-fragment of human IgG2 antibody (centered crystal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-2 chain C region | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Zhao, Y, Gilliland, G. | | Deposit date: | 2012-09-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | IgG2 Fc structure and the dynamic features of the IgG CH2-CH3 interface.
Mol.Immunol., 56, 2013
|
|
3P53
 
 | | Structure of fascin | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, DODECAETHYLENE GLYCOL, Fascin, ... | | Authors: | Jansen, S, Dominguez, R. | | Deposit date: | 2010-10-07 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of actin filament bundling by fascin.
J.Biol.Chem., 286, 2011
|
|
4GOK
 
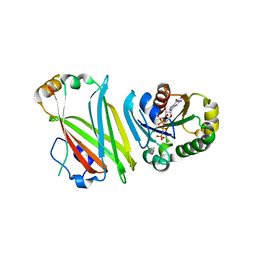 | | The Crystal structure of Arl2GppNHp in complex with UNC119a | | Descriptor: | ADP-ribosylation factor-like protein 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ismail, S, Xiang-Chen, Y, Miertzschke, M, Vetter, I, Koerner, C, Wittinghofer, A. | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119.
Embo J., 31, 2012
|
|
4GPV
 
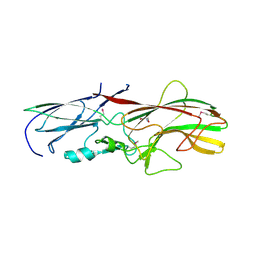 | |
4GOQ
 
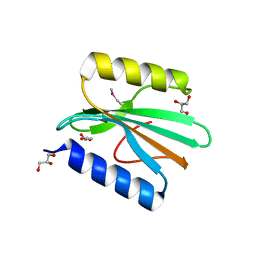 | |
4GXY
 
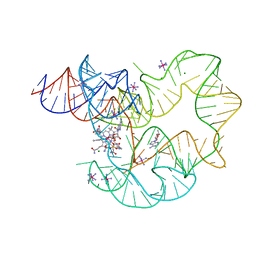 | | RNA structure | | Descriptor: | Adenosylcobalamin, Adenosylcobalamin riboswitch, IRIDIUM HEXAMMINE ION, ... | | Authors: | Serganov, A, Peselis, A. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4H1X
 
 | |
4H5I
 
 | |
