6HTE
 
 | | Sulfolobus solfataricus Tryptophan Synthase B2a | | Descriptor: | DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE, Tryptophan synthase beta chain 2 | | Authors: | Fleming, J, Mayans, O, Bucher, R. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Evolutionary Morphing of Tryptophan Synthase: Functional Mechanisms for the Enzymatic Channeling of Indole.
J.Mol.Biol., 430, 2018
|
|
1ZWL
 
 | |
5EOC
 
 | | Crystal structure of Fab C2 in complex with a Cyclic variant of Hepatitis C Virus E2 epitope I | | Descriptor: | ALA-CYS-GLN-LEU-ILE-ASN-THR-ASN-GLY-SER-TRP-HIS-ILE-CYS, Fab fragment (Heavy chain), Fab fragment (Light chain) | | Authors: | Berisio, R, Ruggiero, A, Sandomenico, A, Patel, A.H, Ruvo, M, Vitagliano, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generation and Characterization of Monoclonal Antibodies against a Cyclic Variant of Hepatitis C Virus E2 Epitope 412-422.
J.Virol., 90, 2016
|
|
1PPC
 
 | | GEOMETRY OF BINDING OF THE BENZAMIDINE-AND ARGININE-BASED INHIBITORS N-ALPHA-(2-NAPHTHYL-SULPHONYL-GLYCYL)-DL-P-AMIDINOPHENYLALANYL-PIPERIDINE (NAPAP) AND (2R,4R)-4-METHYL-1-[N-ALPHA-(3-METHYL-1,2,3,4-TETRAHYDRO-8-QUINOLINESULPHONYL)-L-ARGINYL]-2-PIPERIDINE CARBOXYLIC ACID (MQPA) TO HUMAN ALPHA-THROMBIN: X-RAY CRYSTALLOGRAPHIC DETERMINATION OF THE NAPAP-TRYPSIN COMPLEX AND MODELING OF NAPAP-THROMBIN AND MQPA-THROMBIN | | Descriptor: | 1-[N-(naphthalen-2-ylsulfonyl)glycyl-4-carbamimidoyl-D-phenylalanyl]piperidine, CALCIUM ION, TRYPSIN | | Authors: | Bode, W, Turk, D. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Geometry of binding of the benzamidine- and arginine-based inhibitors N alpha-(2-naphthyl-sulphonyl-glycyl)-DL-p-amidinophenylalanyl-pipe ridine (NAPAP) and (2R,4R)-4-methyl-1-[N alpha-(3-methyl-1,2,3,4-tetrahydr quinolinesulphonyl)-L-arginyl]-2-piperidine carboxylic acid (MQPA) to human alpha-thrombin.X-ray crystallographic determination of the NAPAP-trypsin complex and modeling of NAPAP-thrombin and MQPA-thrombin.
Eur.J.Biochem., 193, 1990
|
|
9OGK
 
 | | Refinement of PDB-3J5R against EMD-8117 using EMAN2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 1 | | Authors: | Chen, M. | | Deposit date: | 2025-04-30 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Building molecular model series from heterogeneous CryoEM structures using Gaussian mixture models and deep neural networks.
Commun Biol, 8, 2025
|
|
2QVJ
 
 | | Crystal structure of a vesicular stomatitis virus nucleocapsid protein Ser290Trp mutant | | Descriptor: | Nucleocapsid protein | | Authors: | Luo, M, Green, T.J, Zhang, X, Tsao, J, Qiu, S. | | Deposit date: | 2007-08-08 | | Release date: | 2008-01-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation.
J.Virol., 82, 2008
|
|
6QKY
 
 | | Tryptophan synthase subunit alpha from Streptococcus pneumoniae with 3D domain swap in the core of TIM barrel | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Michalska, K, Kowiel, M, Bigelow, L, Endres, M, Gilski, M, Jaskolski, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3D domain swapping in the TIM barrel of the alpha subunit of Streptococcus pneumoniae tryptophan synthase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5IXJ
 
 | |
1II4
 
 | | CRYSTAL STRUCTURE OF SER252TRP APERT MUTANT FGF RECEPTOR 2 (FGFR2) IN COMPLEX WITH FGF2 | | Descriptor: | FIBROBLAST GROWTH FACTOR RECEPTOR 2, HEPARIN-BINDING GROWTH FACTOR 2 | | Authors: | Ibrahimi, O.A, Eliseenkova, A.V, Plotnikov, A.N, Ornitz, D.M, Mohammadi, M. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2JIY
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M149 REPLACED WITH TRP (CHAIN M, AM149W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Potter, J.A, Cheng, J, Williams, C.M, Watson, A.J, Jones, M.R. | | Deposit date: | 2007-07-03 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Responses to Cavity-Creating Mutations in an Integral Membrane Protein.
Biochemistry, 46, 2007
|
|
2JJ0
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M248 REPLACED WITH TRP (CHAIN M, AM248W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Fyfe, P.K, Potter, J.A, Cheng, J, Williams, C.M, Watson, A.J, Jones, M.R. | | Deposit date: | 2007-07-03 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Responses to Cavity-Creating Mutations in an Integral Membrane Protein.
Biochemistry, 46, 2007
|
|
1RD5
 
 | | Crystal structure of Tryptophan synthase alpha chain homolog BX1: a member of the chemical plant defense system | | Descriptor: | MALONIC ACID, Tryptophan synthase alpha chain, chloroplast | | Authors: | Kulik, V, Hartmann, E, Weyand, M, Frey, M, Gierl, A, Niks, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2003-11-05 | | Release date: | 2004-12-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes.
J.Mol.Biol., 352, 2005
|
|
1TJR
 
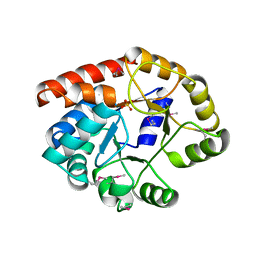 | | Crystal structure of wild-type BX1 complexed with a sulfate ion | | Descriptor: | BX1, SULFATE ION | | Authors: | Kulik, V, Hartmann, E, Weyand, M, Frey, M, Gierl, A, Niks, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2004-06-07 | | Release date: | 2005-08-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes.
J.Mol.Biol., 352, 2005
|
|
8EGZ
 
 | | Engineered tyrosine synthase (TmTyrS1) derived from T. maritima TrpB with Ser bound as the amino-acrylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, POTASSIUM ION, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
8EH0
 
 | | Engineered tyrosine synthase (TmTyrS1) derived from T. maritima TrpB with Ser bound as the amino-acrylate intermediate and complexed with quinoline N-oxide | | Descriptor: | 1,2-ETHANEDIOL, 1-oxo-1lambda~5~-quinoline, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
8EH1
 
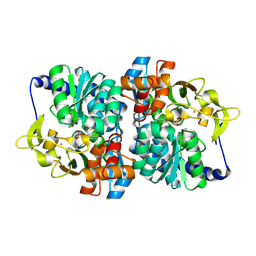 | | Engineered tyrosine synthase (TmTyrS1) derived from T. maritima TrpB with Ser bound as the amino-acrylate intermediate and complexed with 4-hydroxyquinoline | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Engineered tyrosine synthase (TmTyrS1), POTASSIUM ION, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
8EGY
 
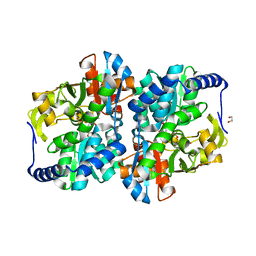 | | Engineered holo tyrosine synthase (TmTyrS1) derived from T. maritima TrpB | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
2G36
 
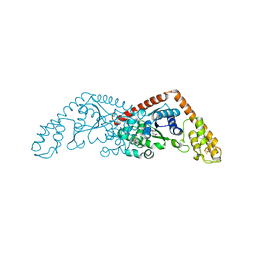 | |
7RNQ
 
 | |
2DH6
 
 | | Crystal structure of E. coli Apo-TrpB | | Descriptor: | SULFATE ION, tryptophan synthase beta subunit | | Authors: | Nishio, K, Morimoto, Y, Ogasahara, K, Yasuoka, N, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding
Febs J., 277, 2010
|
|
2DH5
 
 | | Crystal structure of E. coli Holo-TrpB | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, ... | | Authors: | Nishio, K, Morimoto, Y, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding
Febs J., 277, 2010
|
|
4IPN
 
 | | The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae | | Descriptor: | 6-O-phosphono-alpha-L-idopyranose-(1-4)-4-thio-beta-D-glucopyranose, 6-phospho-beta-glucosidase | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
4IPL
 
 | | The crystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae | | Descriptor: | 6-phospho-beta-glucosidase, GLYCEROL | | Authors: | Yu, W.L, Jiang, Y.L, Andreas, P, Cheng, W, Bai, X.H, Ren, Y.M, Thompsonn, J, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural insights into the substrate specificity of a 6-phospho-&[beta]-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4
J.Biol.Chem., 288, 2013
|
|
1I1Q
 
 | |
1T0W
 
 | | 25 NMR structures of Truncated Hevein of 32 aa (Hevein-32) complex with N,N,N-triacetylglucosamina | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevein | | Authors: | Aboitiz, N, Vila-Perello, M, Groves, P, Asensio, J.L, Andreu, D, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2004-04-13 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR and modeling studies of protein-carbohydrate interactions: synthesis, three-dimensional structure, and recognition properties of a minimum hevein domain with binding affinity for chitooligosaccharides
Chembiochem, 5, 2004
|
|
