3G6L
 
 | |
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | Descriptor: | CAFFEINE, Chitinase | | Authors: | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | Deposit date: | 2009-02-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
3FND
 
 | |
3FXY
 
 | | Acidic Mammalian Chinase, Catalytic Domain | | Descriptor: | Acidic mammalian chitinase | | Authors: | Olland, A.M. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Triad of polar residues implicated in pH specificity of acidic mammalian chitinase.
Protein Sci., 18, 2009
|
|
3FY1
 
 | | The Acidic Mammalian Chitinase catalytic domain in complex with methylallosamidin | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, Acidic mammalian chitinase | | Authors: | Olland, A.M. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triad of polar residues implicated in pH specificity of acidic mammalian chitinase.
Protein Sci., 18, 2009
|
|
3HU7
 
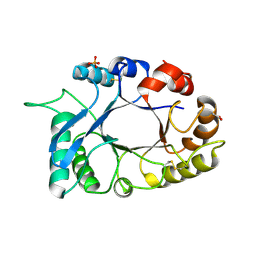 | | Structural characterization and binding studies of a plant pathogenesis related protein heamanthin from haemanthus multiflorus reveal its dual inhibitory effects against xylanase and alpha-amylase | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-06-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
3IAN
 
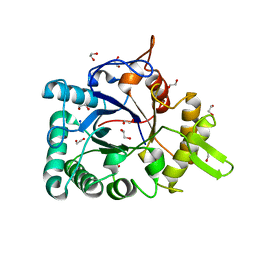 | | Crystal structure of a chitinase from Lactococcus lactis subsp. lactis | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, SODIUM ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a chitinase from Lactococcus lactis subsp. lactis
To be Published
|
|
4W5Z
 
 | |
4URI
 
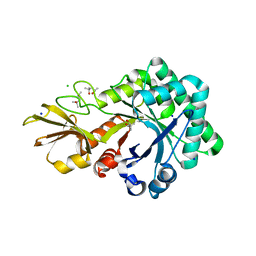 | | Crystal structure of chitinase-like agglutinin RobpsCRA from Robinia pseudoacacia | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHITINASE-RELATED AGGLUTININ, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Henrissat, B, van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2014-06-30 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Carbohydrate Binding Properties of a Plant Chitinase-Like Agglutinin with Conserved Catalytic Machinery.
J.Struct.Biol., 190, 2015
|
|
3OIH
 
 | | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, M, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-19 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution
To be Published
|
|
4WIW
 
 | | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-26 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2
To Be Published
|
|
4WK9
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (0.3mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.102 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1SR0
 
 | | Crystal structure of signalling protein from sheep(SPS-40) at 3.0A resolution using crystal grown in the presence of polysaccharides | | Descriptor: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, signal processing protein | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Singh, N, Kumar, J, Sharma, S, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of signalling protein from sheep(SPS-40) at 3.0A resolution using crystal grown in the presence of polysaccharides
To be Published
|
|
4WJX
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain at 1.0 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R5E
 
 | | Crystal Structure of Family GH18 Chitinase from Cycas revoluta a Complex with Allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ACETATE ION, ALLOSAMIZOLINE, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2014-08-21 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures and inhibitor binding properties of plant class V chitinases: the cycad enzyme exhibits unique structural and functional features.
Plant J., 82, 2015
|
|
4WKH
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (1mM) at 1.05 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKA
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKF
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (2.5mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1TFV
 
 | | CRYSTAL STRUCTURE OF A BUFFALO SIGNALING GLYCOPROTEIN (SPB-40) SECRETED DURING INVOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, mammary gland protein 40 | | Authors: | Bilgrami, S, Saravanan, K, Yadav, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CRYSTAL STRUCTURE OF A BUFFALO SIGNALING GLYCOPROTEIN (SPB-40)
SECRETED DURING INVOLUTION
To be Published
|
|
3QOK
 
 | |
4TOQ
 
 | |
4RL3
 
 | | Crystal Structure of the Catalytic Domain of a family GH18 Chitinase from fern, Peteris ryukyuensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chitinase A, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A class III chitinase without disulfide bonds from the fern, Pteris ryukyuensis: crystal structure and ligand-binding studies.
Planta, 242, 2015
|
|
4TXE
 
 | | ScCTS1 in complex with compound 5 | | Descriptor: | (2S)-1-(2,3-dihydro-1H-inden-2-ylamino)-3-(3,4-dimethylphenoxy)propan-2-ol, Endochitinase | | Authors: | Schuettelkopf, A.W, van Aalten, D.M.F. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors
FEBS Lett, 588, 2014
|
|
4TX6
 
 | | AfChiA1 in complex with compound 1 | | Descriptor: | 3-(2-methoxyphenyl)-6-methyl[1,2]oxazolo[5,4-d]pyrimidin-4(5H)-one, Class III chitinase ChiA1, PHOSPHATE ION | | Authors: | van Aalten, D.M.F. | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors
FEBS Lett, 588, 2014
|
|
3O9N
 
 | | Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Singh, A, Kumar, S, Sinha, M, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution
To be Published
|
|
