3KLH
 
 | | Crystal structure of AZT-Resistant HIV-1 Reverse Transcriptase crosslinked to post-translocation AZTMP-Terminated DNA (COMPLEX P) | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Tu, X, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2IBS
 
 | | Crystal structure of the adenine-specific DNA methyltransferase M.TaqI complexed with the cofactor analog AETA and a 10 bp DNA containing 2-aminopurine at the target position | | Descriptor: | 5'-D(*GP*AP*CP*AP*TP*CP*GP*(6MA)P*AP*C)-3', 5'-D(*GP*TP*TP*CP*GP*(2PR)P*TP*GP*TP*C)-3', 5'-DEOXY-5'-[2-(AMINO)ETHYLTHIO]ADENOSINE, ... | | Authors: | Pljevaljcic, G, Lenz, T, Scheidig, A.J, Weinhold, E. | | Deposit date: | 2006-09-12 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Aminopurine Flipped into the Active Site of the Adenine-Specific DNA Methyltransferase M.TaqI: Crystal Structures and Time-Resolved Fluorescence
J.Am.Chem.Soc., 129, 2007
|
|
2LH1
 
 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | Descriptor: | ACETATE ION, LEGHEMOGLOBIN (ACETO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | Deposit date: | 1982-04-23 | | Release date: | 1983-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
2LH5
 
 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | Descriptor: | FLUORIDE ION, LEGHEMOGLOBIN (FLUORO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | Deposit date: | 1982-04-23 | | Release date: | 1983-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
2LH3
 
 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | Descriptor: | CYANIDE ION, LEGHEMOGLOBIN (CYANO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | Deposit date: | 1982-04-23 | | Release date: | 1983-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
8TNR
 
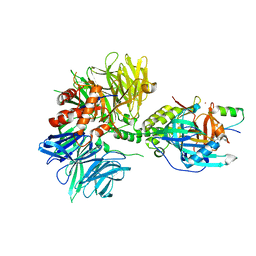 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
7SEP
 
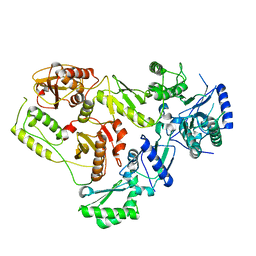 | |
2SNS
 
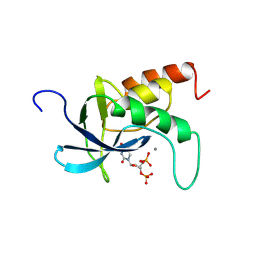 | | STAPHYLOCOCCAL NUCLEASE. PROPOSED MECHANISM OF ACTION BASED ON STRUCTURE OF ENZYME-THYMIDINE 3(PRIME),5(PRIME)-BIPHOSPHATE-CALCIUM ION COMPLEX AT 1.5-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, THERMONUCLEASE PRECURSOR, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Legg, M.J, Cotton, F.A, Hazen Jr, E.E. | | Deposit date: | 1982-05-14 | | Release date: | 1982-07-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
Thesis, Texas Agricultural and Mechanical University, 1977
|
|
8TUR
 
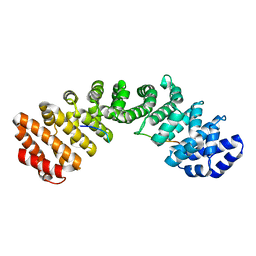 | |
8WKL
 
 | |
6XXG
 
 | | Structure of truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Deinococcus radiodurans | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-01-27 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Identification of a 1-deoxy-D-xylulose-5-phosphate synthase (DXS) mutant with improved crystallographic properties.
Biochem.Biophys.Res.Commun., 539, 2021
|
|
8A3G
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil homotetramer with 4 heptad repeats, apCC-Tet* | | Descriptor: | ACETATE ION, SODIUM ION, apCC-Tet* | | Authors: | Naudin, E.A, Mylemans, B, Albanese, K.I, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
7AAC
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with veliparib | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
8W88
 
 | | Cryo-EM structure of the SEP363856-bound TAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8UWB
 
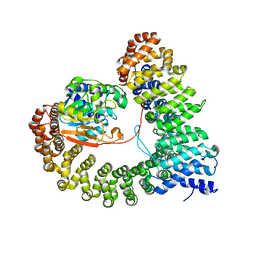 | | Crystal structure of PP2A PPP2R1A-PPP2CA-PPP2R5E phosphatase. | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit epsilon isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wachter, F, Nowak, R.P, Fischer, E.S. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of methylation-independent PP2A assembly guides alphafold2Multimer prediction of family-wide PP2A complexes.
J.Biol.Chem., 300, 2024
|
|
4X89
 
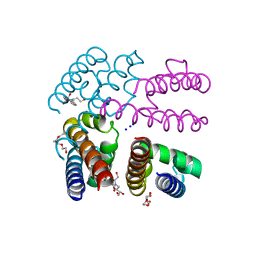 | | NavMs voltage-gated sodium channal pore and C-terminal domain soaked with Silver nitrate | | Descriptor: | HEGA-10, Ion transport protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
8W8B
 
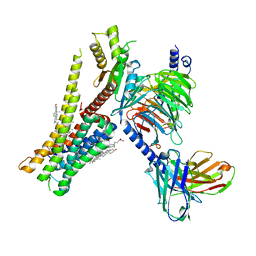 | | Cryo-EM structure of SEP-363856 bounded serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8TLS
 
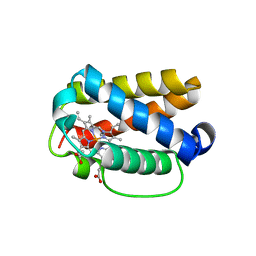 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y108A variant | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schultz, T.D, Martinez, J.E, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y108A variant
To Be Published
|
|
8W87
 
 | | Cryo-EM structure of the METH-TAAR1 complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W8A
 
 | | Cryo-EM structure of the RO5256390-TAAR1 complex | | Descriptor: | (4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-imine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
5LBD
 
 | | Crystal structure of the N-domain of HMA6, a copper-transporting P-type ATPase | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Mayerhofer, H, Ravaud, S, Pebay-Peyroula, E. | | Deposit date: | 2016-06-15 | | Release date: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Nucleotide-Binding Domains of the P1B-type ATPases HMA6 and HMA8 from Arabidopsis thaliana.
PLoS ONE, 11, 2016
|
|
8U3Z
 
 | | Model of TTLL6 bound to microtubule from composite map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-09-08 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 2024
|
|
8UMU
 
 | | Atomic model of the human CTF18-RFC-PCNA binary complex in the four-subunit binding state (state 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8YRU
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (apo form) after 15 sec of soaking with phenylhydrazine | | Descriptor: | ACETATE ION, Aminotransferase class IV, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (apo form) after 15 sec of soaking with phenylhydrazine
To Be Published
|
|
7AAD
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
