7NVX
 
 | | TFIIH in a post-translocated state (with ADP-BeF3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CDK-activating kinase assembly factor MAT1, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NVW
 
 | | TFIIH in a pre-translocated state (without ADP-BeF3) | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription and DNA repair factor IIH helicase subunit XPB, General transcription factor IIE subunit 1, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
4UG0
 
 | | STRUCTURE OF THE HUMAN 80S RIBOSOME | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S RIBOSOMAL PROTEIN, ... | | Authors: | Khatter, H, Myasnikov, A.G, Natchiar, S.K, Klaholz, B.P. | | Deposit date: | 2015-03-20 | | Release date: | 2015-06-10 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the human 80S ribosome
NATURE, 520, 2015
|
|
3D1B
 
 | | Tetragonal crystal structure of Tas3 C-terminal alpha motif | | Descriptor: | RNA-induced transcriptional silencing complex protein tas3 | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.
Mol.Cell, 34, 2009
|
|
4KHP
 
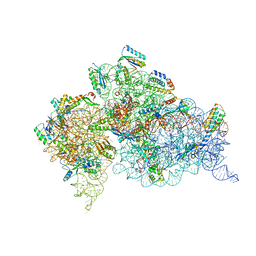 | | Structure of the Thermus thermophilus 30S ribosomal subunit in complex with de-6-MSA-pactamycin | | Descriptor: | 16S Ribosomal RNA, 30S Ribosomal protein S10, 30S Ribosomal protein S11, ... | | Authors: | Tourigny, D.S, Fernandez, I.S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2013-05-01 | | Release date: | 2013-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Bioactive Pactamycin Analog Bound to the 30S Ribosomal Subunit.
J.Mol.Biol., 425, 2013
|
|
1J85
 
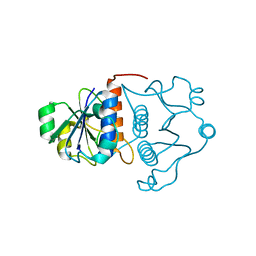 | | Structure of YibK from Haemophilus influenzae (HI0766), a truncated sequence homolog of tRNA (guanosine-2'-O-) methyltransferase (SpoU) | | Descriptor: | YibK | | Authors: | Lim, K, Zhang, H, Toedt, J, Tempcyzk, A, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae
(HI0766): A cofactor bound at a site formed by a knot
Proteins, 51, 2003
|
|
3CQS
 
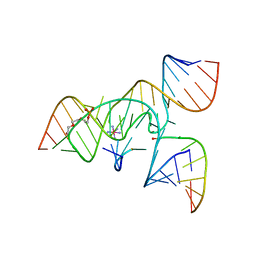 | | A 3'-OH, 2',5'-phosphodiester substitution in the hairpin ribozyme active site reveals similarities with protein ribonucleases | | Descriptor: | 13-mer substrate strand with 3'-OH, 2',5'-phosphodiester covalently linking 5th and 6th nucleotides, 19-mer ribozyme strand, ... | | Authors: | Torelli, A.T, Spitale, R.C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Shared traits on the reaction coordinates of ribonuclease and an RNA enzyme
Biochem.Biophys.Res.Commun., 371, 2008
|
|
4KGN
 
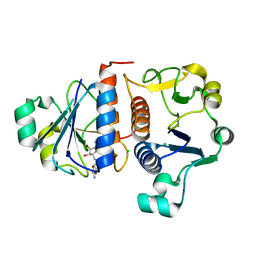 | |
7ASE
 
 | | 43S preinitiation complex from Trypanosoma cruzi with the kDDX60 helicase | | Descriptor: | 18S, 40S ribosomal protein S10, putative, ... | | Authors: | Bochler, A, Brito Querido, J, Prilepskaja, T, Soufari, H, Del Cistia, M.L, Kuhn, L, Rimoldi Ribeiro, A, Valasek, L.S, Hashem, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural Differences in Translation Initiation between Pathogenic Trypanosomatids and Their Mammalian Hosts.
Cell Rep, 33, 2020
|
|
4WWU
 
 | | Structure of Mex67:Mtr2 | | Descriptor: | ZINC ION, mRNA export factor MEX67, mRNA transport regulator MTR2 | | Authors: | Aibara, S, Valkov, E, Stewart, M. | | Deposit date: | 2014-11-12 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Domain organization within the nuclear export factor Mex67:Mtr2 generates an extended mRNA binding surface.
Nucleic Acids Res., 43, 2015
|
|
4WP2
 
 | | Chaetomium Mex67 UBA domain | | Descriptor: | Putative mRNA export protein | | Authors: | Aibara, S, Stewart, M. | | Deposit date: | 2014-10-17 | | Release date: | 2015-07-15 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of the principal mRNA-export factor Mex67-Mtr2 from Chaetomium thermophilum.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3MB5
 
 | |
6QIE
 
 | | Crystal structure of DEAH-box ATPase Prp43-S387G | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Hamann, F, Ficner, R, Enders, M. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for RNA translocation by DEAH-box ATPases.
Nucleic Acids Res., 47, 2019
|
|
1J7T
 
 | |
6QLB
 
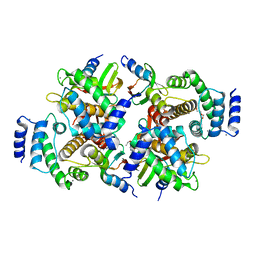 | | Calpain small subunit 1, RNA-binding protein Hfq | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, CALCIUM ION, ... | | Authors: | Rizkallah, P.J, Cresser-Brown, J.O. | | Deposit date: | 2019-01-31 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Calpain small subunit 1, RNA-binding protein Hfq
Acta Crystallogr.,Sect.F, 2020
|
|
6QID
 
 | | Crystal structure of DEAH-box ATPase Prp43-S387A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Hamann, F, Ficner, R, Enders, M. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis for RNA translocation by DEAH-box ATPases.
Nucleic Acids Res., 47, 2019
|
|
7NVV
 
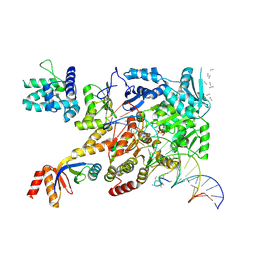 | | XPB-containing part of TFIIH in a post-translocated state (with ADP-BeF3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
2RUG
 
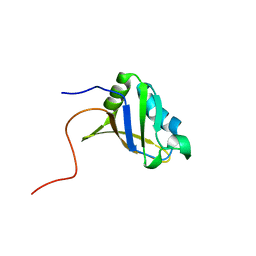 | | Refined solution structure of the first RNA recognition motif domain in CPEB3 | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Tsuda, K, Kuwasako, K, Nagata, T, Takahashi, M, Kigawa, T, Kobayashi, N, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel RNA recognition motif domain in the cytoplasmic polyadenylation element binding protein 3.
Proteins, 82, 2014
|
|
6ZQC
 
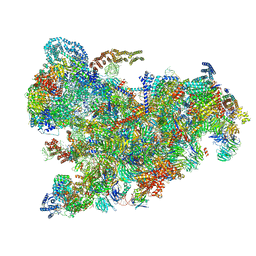 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Pre-A1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQB
 
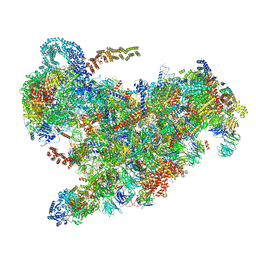 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state B2 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
4K51
 
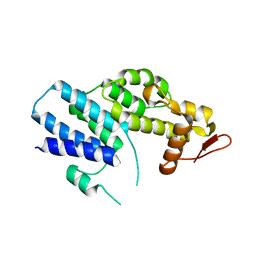 | | Crystal Structure of the PCI domain of eIF3a | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2013-04-12 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural integrity of the PCI domain of eIF3a/TIF32 is required for mRNA recruitment to the 43S pre-initiation complexes.
Nucleic Acids Res., 42, 2014
|
|
4V6C
 
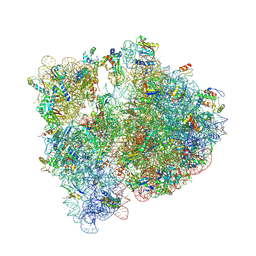 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
6LQS
 
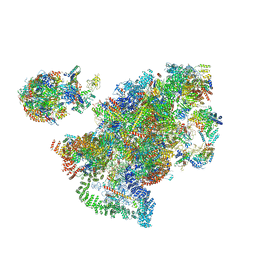 | |
6LQR
 
 | |
6LQV
 
 | |
