3V8C
 
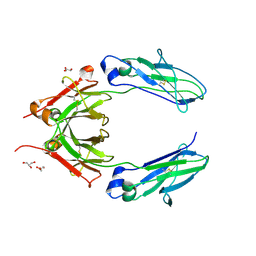 | | Crystal structure of monoclonal human anti-rhesus D Fc IgG1 t125(yb2/0) double mutant (H310 and H435 in K) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, GLYCEROL, ... | | Authors: | Menez, R, Stura, E.A, Bourel, D, Siberil, S, Jorieux, S, De Romeuf, C, Ducancel, F, Fridman, W.H, Teillaud, J.L. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Effect of zinc on human IgG1 and its Fc gamma R interactions.
Immunol.Lett., 143, 2012
|
|
4ZUP
 
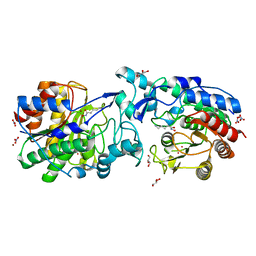 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a hydroxamate inhibitor | | Descriptor: | 5-amino-N-hydroxypentanamide, Acetylpolyamine aminohydrolase, GLYCEROL, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2015-05-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
1YQG
 
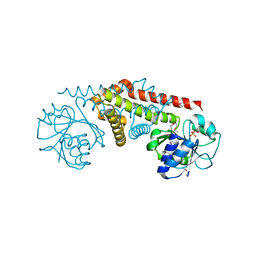 | | Crystal structure of a pyrroline-5-carboxylate reductase from neisseria meningitides mc58 | | Descriptor: | SULFATE ION, pyrroline-5-carboxylate reductase | | Authors: | Chang, C, Joachimiak, A, Li, H, Collart, F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-01 | | Release date: | 2005-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Delta(1)-Pyrroline-5-carboxylate Reductase from Human Pathogens Neisseria meningitides and Streptococcus pyogenes
J.Mol.Biol., 354, 2005
|
|
3V95
 
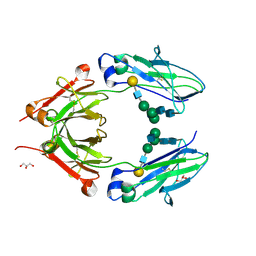 | | Crystal structure of monoclonal human anti-rhesus D Fc and IgG1 t125(yb2/0) in the presence of EDTA | | Descriptor: | GLYCEROL, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Menez, R, A Stura, E, Bourel, D, Siberil, S, Jorieux, S, De Romeuf, C, Ducancel, F, Fridman, W.H, Teillaud, J.L. | | Deposit date: | 2011-12-23 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Effect of zinc on human IgG1 and its Fc gamma R interactions.
Immunol.Lett., 143, 2012
|
|
5OWL
 
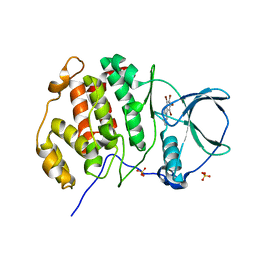 | | Low salt structure of human protein kinase CK2alpha in complex with 3-aminopropyl-4,5,6,7-tetrabromobenzimidazol | | Descriptor: | 3-[4,5,6,7-tetrakis(bromanyl)benzimidazol-1-yl]propan-1-amine, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Bretner, M, Chojnacki, C. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Biological properties and structural study of new aminoalkyl derivatives of benzimidazole and benzotriazole, dual inhibitors of CK2 and PIM1 kinases.
Bioorg. Chem., 80, 2018
|
|
3LRI
 
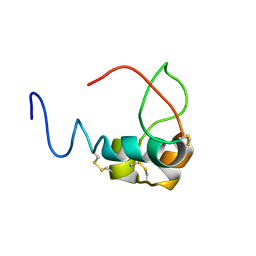 | | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I | | Descriptor: | PROTEIN (INSULIN-LIKE GROWTH FACTOR I) | | Authors: | Laajoki, L.G, Francis, G.L, Wallace, J.C, Carver, J.A, Keniry, M.A. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I
J.Biol.Chem., 275, 2000
|
|
8EC6
 
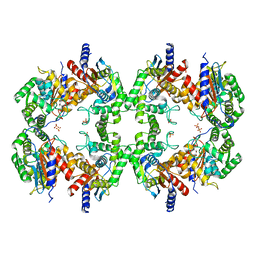 | | Cryo-EM structure of the Glutaminase C core filament (fGAC) | | Descriptor: | Isoform 2 of Glutaminase kidney isoform, mitochondrial, PHOSPHATE ION | | Authors: | Ambrosio, A.L, Dias, S.M, Quesnay, J.E, Portugal, R.V, Cassago, A, van Heel, M.G, Islam, Z, Rodrigues, C.T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of glutaminase activation through filamentation and the role of filaments in mitophagy protection.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6NJZ
 
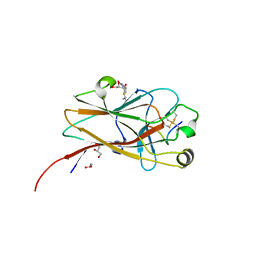 | |
4ZXA
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD in complex with Cd2+ and 4-hydroxybenzonitrile | | Descriptor: | 4-hydroxybenzonitrile, CADMIUM ION, Hydroquinone dioxygenase large subunit, ... | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
1CP5
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN L104F MUTANT (MET) | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Liong, E.C, Phillips Jr, G.N. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Waterproofing the heme pocket. Role of proximal amino acid side chains in preventing hemin loss from myoglobin.
J.Biol.Chem., 276, 2001
|
|
7AQ1
 
 | | Crystal structure of human mature meprin beta in complex with the specific inhibitor MWT-S-270 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Linnert, M, Parthier, C, Fritz, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure and Dynamics of Meprin beta in Complex with a Hydroxamate-Based Inhibitor.
Int J Mol Sci, 22, 2021
|
|
2Y9T
 
 | |
2M1M
 
 | | Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response | | Descriptor: | Auxin-induced protein IAA4 | | Authors: | Kovermann, M, Dinesh, D.C, Gopalswamy, M, Abel, S, Balbach, J. | | Deposit date: | 2012-12-03 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5RPH
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen A11a | | Descriptor: | 3-[3,4-bis(fluoranyl)phenyl]-1,4,6,7-tetrahydroimidazo[2,1-c][1,2,4]triazine, Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1H8B
 
 | | EF-hands 3,4 from alpha-actinin / Z-repeat 7 from titin | | Descriptor: | ALPHA-ACTININ 2, SKELETAL MUSCLE ISOFORM, TITIN | | Authors: | Atkinson, R.A, Joseph, C, Kelly, G, Muskett, F.W, Frenkiel, T.A, Nietlispach, D, Pastore, A. | | Deposit date: | 2001-02-01 | | Release date: | 2001-08-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ca2+-Independent Binding of an EF-Hand Domain to a Novel Motif in the Alpha-Actinin-Titin Complex
Nat.Struct.Biol., 8, 2001
|
|
6NK1
 
 | |
4HA0
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant R230D with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
2OFP
 
 | | Crystal structure of Escherichia coli ketopantoate reductase in a ternary complex with NADP+ and pantoate | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETATE ION, Ketopantoate reductase, ... | | Authors: | Ciulli, A, Chirgadze, D.Y, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli Ketopantoate Reductase in a Ternary Complex with NADP+ and Pantoate Bound: SUBSTRATE RECOGNITION, CONFORMATIONAL CHANGE, AND COOPERATIVITY.
J.Biol.Chem., 282, 2007
|
|
1ZBX
 
 | | Crystal structure of a Orc1p-Sir1p complex | | Descriptor: | Origin recognition complex subunit 1, Regulatory protein SIR1 | | Authors: | Hsu, H.C, Stillman, B, Xu, R.M. | | Deposit date: | 2005-04-09 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1COA
 
 | | THE EFFECT OF CAVITY CREATING MUTATIONS IN THE HYDROPHOBIC CORE OF CHYMOTRYPSIN INHIBITOR 2 | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Jackson, S.E, Moracci, M, Elmasry, N, Johnson, C.M, Fersht, A.R. | | Deposit date: | 1993-05-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2.
Biochemistry, 32, 1993
|
|
8BWL
 
 | | Crystal structure of human Twisted gastrulation protein homolog 1 (TWSG1) in complex with human Growth Differentiation factor 5 (GDF5) and calcium | | Descriptor: | CALCIUM ION, Growth/differentiation factor 5, Twisted gastrulation protein homolog 1 | | Authors: | Malinauskas, T, Rudolf, A.F, Moore, G, Eggington, H, Belnoue-Davis, H, El Omari, K, Woolley, R.E, Griffiths, S.C, Duman, R, Wagner, A, Leedham, S.J, Baldock, C, Ashe, H, Siebold, C. | | Deposit date: | 2022-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular mechanism of BMP signal control by Twisted gastrulation.
Nat Commun, 15, 2024
|
|
8BWM
 
 | | Crystal structure of human Twisted gastrulation protein homolog 1 (TWSG1) in complex with human Growth Differentiation factor 5 (GDF5) and calcium, long-wavelength X-ray dataset (4042 eV) | | Descriptor: | CALCIUM ION, Growth/differentiation factor 5, Twisted gastrulation protein homolog 1 | | Authors: | Malinauskas, T, Rudolf, A.F, Moore, G, Eggington, H, Belnoue-Davis, H, El Omari, K, Woolley, R.E, Griffiths, S.C, Duman, R, Wagner, A, Leedham, S.J, Baldock, C, Ashe, H, Siebold, C. | | Deposit date: | 2022-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of BMP signal control by Twisted gastrulation.
Nat Commun, 15, 2024
|
|
7DS2
 
 | |
8BWN
 
 | | Crystal structure of human Twisted gastrulation protein homolog 1 (TWSG1) in complex with human Growth Differentiation Factor 5 (GDF5) and calcium, long-wavelength X-ray dataset (4010 eV) | | Descriptor: | CALCIUM ION, Growth/differentiation factor 5, Twisted gastrulation protein homolog 1 | | Authors: | Malinauskas, T, Rudolf, A.F, Moore, G, Eggington, H, Belnoue-Davis, H, El Omari, K, Woolley, R.E, Griffiths, S.C, Duman, R, Wagner, A, Leedham, S.J, Baldock, C, Ashe, H, Siebold, C. | | Deposit date: | 2022-12-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Molecular mechanism of BMP signal control by Twisted gastrulation.
Nat Commun, 15, 2024
|
|
7DSB
 
 | |
