3VD2
 
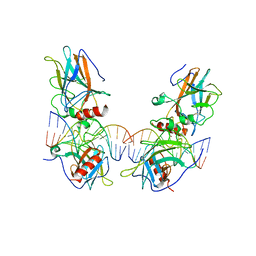 | | structure of p73 DNA binding domain tetramer modulates p73 transactivation | | Descriptor: | DNA (5'-D(*AP*TP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*AP*T)-3'), Tumor protein p73, ZINC ION | | Authors: | Ethayathulla, A.S, Tse, P.W, Nguyen, S, Viadiu, H. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of p73 DNA-binding domain tetramer modulates p73 transactivation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3W11
 
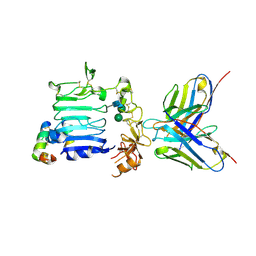 | |
8C08
 
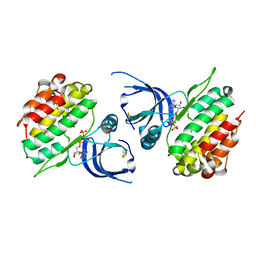 | | Crystal structure of JAK2 JH2-K539L | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Haikarainen, T. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of JAK2 activation in erythropoietin receptor and pathogenic JAK2 signaling.
Sci Adv, 10, 2024
|
|
2RTA
 
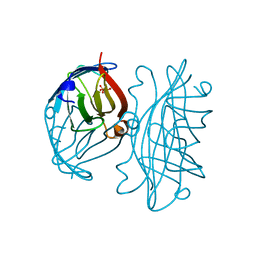 | | APOSTREPTAVIDIN, PH 2.97, SPACE GROUP I4122 | | Descriptor: | STREPTAVIDIN, SULFATE ION | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
8C0A
 
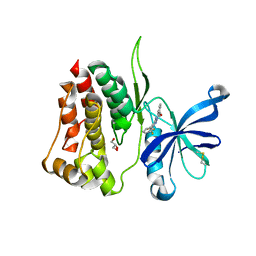 | | Crystal structure of JAK2 JH2-R683S | | Descriptor: | 3,5-diphenyl-2-(trifluoromethyl)-1~{H}-pyrazolo[1,5-a]pyrimidin-7-one, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of JAK2 activation in erythropoietin receptor and pathogenic JAK2 signaling.
Sci Adv, 10, 2024
|
|
7OL2
 
 | | Crystal structure of mouse contactin 1 immunoglobulin domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, ... | | Authors: | Chataigner, L.M.P, Janssen, B.J.C. | | Deposit date: | 2021-05-19 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structural insights into the contactin 1 - neurofascin 155 adhesion complex.
Nat Commun, 13, 2022
|
|
7OL4
 
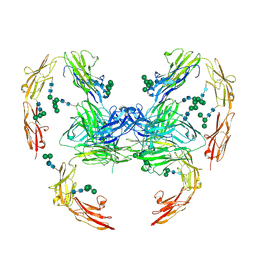 | |
8C09
 
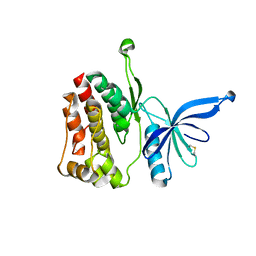 | | Crystal structure of JAK2 JH2-I559F | | Descriptor: | Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of JAK2 activation in erythropoietin receptor and pathogenic JAK2 signaling.
Sci Adv, 10, 2024
|
|
2RTN
 
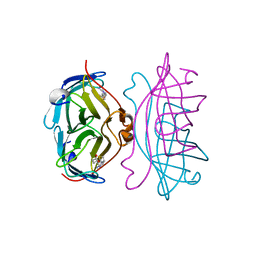 | | STREPTAVIDIN-2-IMINOBIOTIN COMPLEX, PH 2.0, SPACE GROUP I222 | | Descriptor: | 2-IMINOBIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
6E6L
 
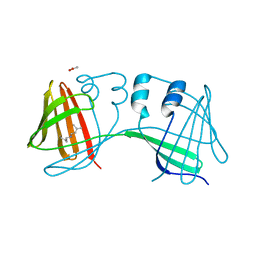 | |
8CON
 
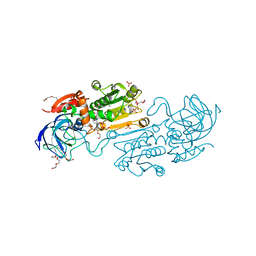 | | Crystal structure of alcohol dehydrogenase from Arabidopsis thaliana in complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase class-P, ... | | Authors: | Fermani, S, Fanti, S, Carloni, G, Falini, G, Meloni, M, Zaffagnini, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of Arabidopsis alcohol dehydrogenases reveals distinct functional properties but similar redox sensitivity.
Plant J., 118, 2024
|
|
7PQD
 
 | | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 A: the structural basis for dimerisation | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Qian, P, Hunter, C.N. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the dimeric Rhodobacter sphaeroides RC-LH1 core complex at 2.9 angstrom : the structural basis for dimerisation.
Biochem.J., 478, 2021
|
|
3W13
 
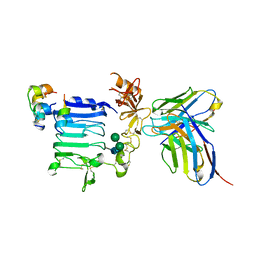 | | Insulin receptor ectodomain construct comprising domains L1-CR in complex with high-affinity insulin analogue [D-PRO-B26]-DTI-NH2, alphact peptide(693-719) and FAB 83-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Lawrence, M.C, Smith, B.J, Brzozowski, A.M. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.303 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor
Nature, 493, 2013
|
|
6EAT
 
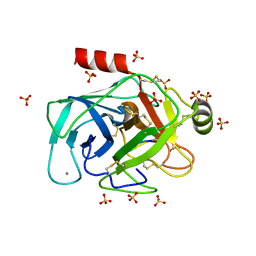 | | Crystallographic structure of the cyclic nonapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21. | | Descriptor: | 9MER-PEPTIDE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Crystallographic structure of a complex between trypsin and a nonapeptide derived from a Bowman-Birk inhibitor found in Vigna unguiculata seeds.
Arch. Biochem. Biophys., 665, 2019
|
|
2M49
 
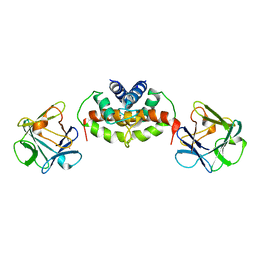 | |
7PTU
 
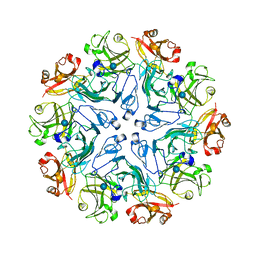 | |
6F2A
 
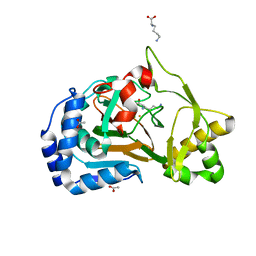 | | Crystal structure of the complex Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO1 with Fe(II)/Lysine | | Descriptor: | ACETIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Isabet, T, Stura, E.A, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-11-24 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
6F1D
 
 | | CUB2 domain of C1r | | Descriptor: | CALCIUM ION, Complement C1r subcomponent, SODIUM ION | | Authors: | Almitairi, J.O.M, Venkatraman Girija, U, Furze, C.M, Simpson-Gray, X, Badakshi, F, Marshall, J.E, Mitchell, D.A, Moody, P.C.E, Wallis, R. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the C1r-C1s interaction of the C1 complex of complement activation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6F2O
 
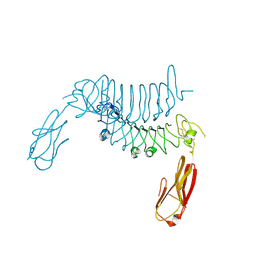 | |
3W12
 
 | | Insulin receptor ectodomain construct comprising domains L1-CR in complex with high-affinity insulin analogue [D-PRO-B26]-DTI-NH2, alpha-CT peptide(704-719) and FAB 83-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Lawrence, M.C, Smith, B.J, Brzozowsk, A.M. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.301 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor
Nature, 493, 2013
|
|
1JC6
 
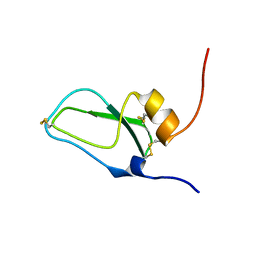 | | SOLUTION STRUCTURE OF BUNGARUS FACIATUS IX, A KUNITZ-TYPE CHYMOTRYPSIN INHIBITOR | | Descriptor: | VENOM BASIC PROTEASE INHIBITORS IX AND VIIIB | | Authors: | Chen, C, Hsu, C.H, Su, N.Y, Chiou, S.H, Wu, S.H. | | Deposit date: | 2001-06-08 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Kunitz-type chymotrypsin inhibitor isolated from the elapid snake Bungarus fasciatus
J.BIOL.CHEM., 276, 2001
|
|
6F9G
 
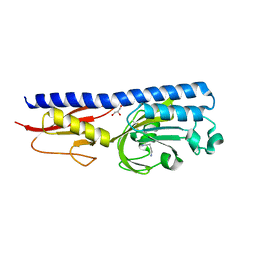 | | Ligand binding domain of P. putida KT2440 polyamine chemorecpetors McpU in complex putrescine. | | Descriptor: | 1,4-DIAMINOBUTANE, ACETATE ION, GLYCEROL, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M.T, Ortega, A, Martin-Mora, D, Corral-Lugo, A, Morel, B, Krell, T. | | Deposit date: | 2017-12-14 | | Release date: | 2018-03-28 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structural Basis for Polyamine Binding at the dCACHE Domain of the McpU Chemoreceptor from Pseudomonas putida.
J. Mol. Biol., 430, 2018
|
|
5NYW
 
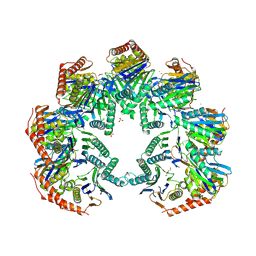 | | Anbu (ancestral beta-subunit) from Yersinia bercovieri | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Piasecka, A, Czapinska, H, Vielberg, M, Szczepanowski, R.H, Reed, S, Groll, M, Bochtler, M. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | The Y. bercovieri Anbu crystal structure sheds light on the evolution of highly (pseudo)symmetric multimers.
J. Mol. Biol., 430, 2018
|
|
6ETL
 
 | |
6F2B
 
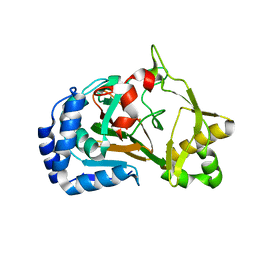 | | Crystal structure of the complex Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO1 with Fe(II)/alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, L-lysine 3-hydroxylase | | Authors: | Isabet, T, Stura, E.A, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-11-24 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
