5A4A
 
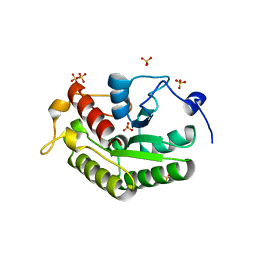 | | Crystal structure of the OSK domain of Drosophila Oskar | | Descriptor: | MATERNAL EFFECT PROTEIN OSKAR, SULFATE ION | | Authors: | Jeske, M, Glatt, S, Ephrussi, A, Mueller, C.W. | | Deposit date: | 2015-06-05 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | The Crystal Structure of the Drosophila Germline Inducer Oskar Identifies Two Domains with Distinct Vasa Helicase-and RNA-Binding Activities.
Cell Rep., 12, 2015
|
|
8Y0E
 
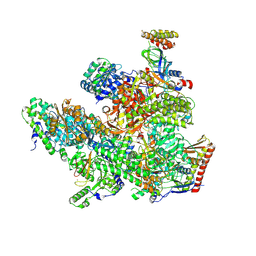 | | ASFV RNAP M1249L C-tail occupied complex4 (MCOC4) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-22 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
5VPN
 
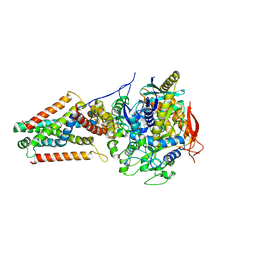 | | E. coli Quinol fumarate reductase FrdA E245Q mutation | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Starbird, C.A, Maklashina, E, Sharma, P, Qualls-Histed, S, Cecchini, G, Iverson, T.M. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2232 Å) | | Cite: | Structural and biochemical analyses reveal insights into covalent flavinylation of the Escherichia coli Complex II homolog quinol:fumarate reductase.
J. Biol. Chem., 292, 2017
|
|
383D
 
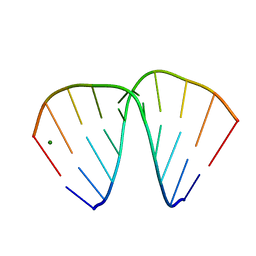 | |
4J8S
 
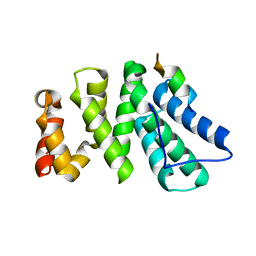 | | Crystal structure of human CNOT1 MIF4G domain in complex with a TTP peptide | | Descriptor: | CCR4-NOT transcription complex subunit 1, Tristetraprolin | | Authors: | Frank, F, Fabian, M.R, Rouya, C, Siddiqui, N, Lai, W.S, Karetnikov, A, Blackshear, P.J, Sonenberg, N, Nagar, B. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2LJZ
 
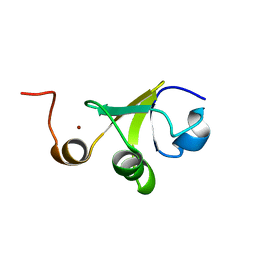 | | Structure of the C-terminal domain of HPV16 E6 oncoprotein | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
5KQF
 
 | | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-4-methyl-6-pyrimidin-5-yl-5,6-dihydro-1,3-thiazin-2-amine (compound 12) bound to BACE1 | | Descriptor: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-4-methyl-6-pyrimidin-5-yl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1 | | Authors: | Lewis, H.A, Wu, Y.J, Rajamani, R, Thompson, L.A. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of S3-Truncated, C-6 Heteroaryl Substituted Aminothiazine beta-Site APP Cleaving Enzyme-1 (BACE1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
8XX5
 
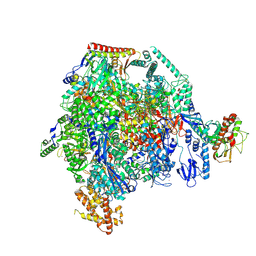 | | ASFV RNAP M1249L C-tail occupied complex1 (MCOC1) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
5KR8
 
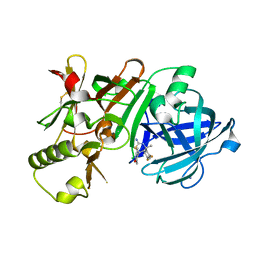 | | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine (compound 5) bound to BACE1 | | Descriptor: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1, IODIDE ION | | Authors: | Lewis, H.A, Wu, Y.J, Rajamani, R, Thompson, L.A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Discovery of S3-Truncated, C-6 Heteroaryl Substituted Aminothiazine beta-Site APP Cleaving Enzyme-1 (BACE1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
3LJA
 
 | |
4XEY
 
 | |
8XY6
 
 | | ASFV RNAP M1249L C-tail occupied complex3 (MCOC3) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
5KVD
 
 | | Zika specific antibody, ZV-2, bound to ZIKA envelope DIII | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
1E3A
 
 | | A slow processing precursor penicillin acylase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hewitt, L, Kasche, V, Lummer, K, Lewis, R.J, Murshudov, G.N, Verma, C.S, Dodson, G.G, Wilson, K.S. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Slow Processing Precursor Penicillin Acylase from Escherichia Coli Reveals the Linker Peptide Blocking the Active-Site Cleft
J.Mol.Biol., 302, 2000
|
|
7WID
 
 | | Crystal structure of Staphylococcus aureus ClpP in complex with ZG180 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wei, B.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2022-01-03 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
6ZTC
 
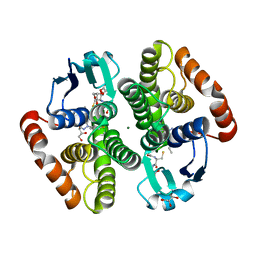 | | CRYSTAL STRUCTURE OF PROSTAGLANDIN D2 SYNTHASE IN COMPLEX WITH FRAGMENT 1A AT 1.84A RESOLUTION. | | Descriptor: | 1-[1-(3-fluorophenyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]propan-1-one, GLUTATHIONE, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
5O3X
 
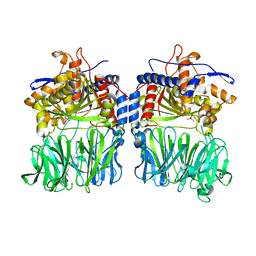 | | Structural characterization of the fast and promiscuous macrocyclase from plant - apo PCY1 | | Descriptor: | CACODYLATE ION, Peptide cyclase 1 | | Authors: | Ludewig, H, Czekster, C.M, Bent, A.F, Naismith, J.H. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.
ACS Chem. Biol., 13, 2018
|
|
5AWB
 
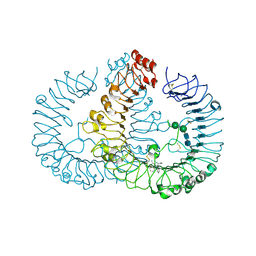 | | Crystal structure of human TLR8 in complex with N1-3-aminomethylbenzyl (meta-amine) | | Descriptor: | 1-[[3-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Human TLR8-Specific Agonists with Augmented Potency and Adjuvanticity.
J.Med.Chem., 58, 2015
|
|
5NOF
 
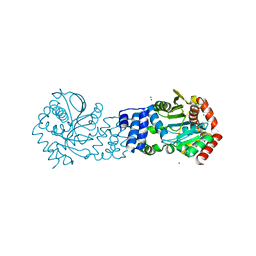 | | Anthranilate phosphoribosyltransferase from Thermococcus kodakaraensis | | Descriptor: | Anthranilate phosphoribosyltransferase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Perveen, S, Rashid, N, Papageorgiou, A.C. | | Deposit date: | 2017-04-12 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Anthranilate phosphoribosyltransferase from the hyperthermophilic archaeon Thermococcus kodakarensis shows maximum activity with zinc and forms a unique dimeric structure.
FEBS Open Bio, 7, 2017
|
|
9HC2
 
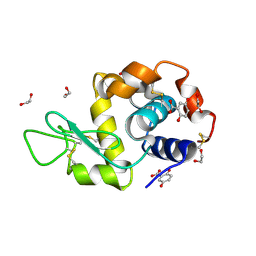 | | Crystal structure of Lysozyme in complex with Gentisic Acid | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dihydroxybenzoic acid, Lysozyme C | | Authors: | Ifeagwu, M.C, Flood, R.J, Mockler, N.M, Crowley, P.B. | | Deposit date: | 2024-11-08 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of Lysozyme in complex with Gentisic Acid
To Be Published
|
|
9CIF
 
 | |
8CBR
 
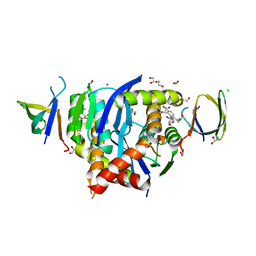 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BDM-2 | | Descriptor: | (2S)-2-[3-cyclopropyl-2-(3,4-dihydro-2H-chromen-6-yl)-6-methyl-phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBV
 
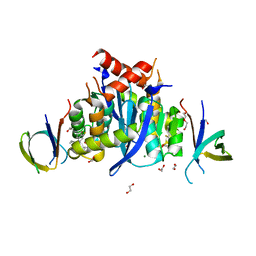 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT916 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-2-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-6-methyl-phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
7WGS
 
 | | Structure of ClpP from Staphylococcus aureus in complex with (S)-ZG197 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6S,9aS)-6-[(2R)-butan-2-yl]-8-[(1S)-1-naphthalen-1-ylethyl]-4,7-bis(oxidanylidene)-N-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Yang, C.-G, Gan, J.H. | | Deposit date: | 2021-12-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP.
Nat Commun, 13, 2022
|
|
6JIB
 
 | | Human MTHFD2 in complex with DS44960156 | | Descriptor: | 4-(5-oxo-1,5-dihydro-2H-[1]benzopyrano[3,4-c]pyridine-3(4H)-carbonyl)benzoic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Suzuki, M, Matsui, Y, Kawai, J. | | Deposit date: | 2019-02-20 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design and Synthesis of an Isozyme-Selective MTHFD2 Inhibitor with a Tricyclic Coumarin Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
