4YRU
 
 | | Crystal structure of C-terminally truncated Neuronal Calcium Sensor (NCS-1) from Rattus norvegicus | | Descriptor: | CALCIUM ION, Neuronal calcium sensor 1 | | Authors: | Pandalaneni, S, Karrupiah, V, Mayans, O, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-03-15 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-protein-coupled Receptor Kinase 1 (GRK1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
4E8C
 
 | | Crystal structure of streptococcal beta-galactosidase in complex with galactose | | Descriptor: | GLYCEROL, Glycosyl hydrolase, family 35, ... | | Authors: | Cheng, W, Wang, L, Bai, X.H, Jiang, Y.L, Li, Q, Yu, G, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
J.Biol.Chem., 287, 2012
|
|
7VQN
 
 | | Crystal structure of KPC-2 beta-lactamase complexed with hydrolyzed EXW-1 | | Descriptor: | (2R,3S)-3-methyl-4-methylidene-2-[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-2,3-dihydropyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, SULFATE ION | | Authors: | Xie, H.X. | | Deposit date: | 2021-10-20 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of KPC-2 beta-lactamase complexed with hydrolyzed EXW-1
To Be Published
|
|
4EGR
 
 | | 2.50 angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with phosphoenolpyruvate | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, PHOSPHOENOLPYRUVATE, SULFATE ION | | Authors: | Krishna, S.N, Light, S.H, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.50 angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with phosphoenolpyruvate
TO BE PUBLISHED
|
|
7VMA
 
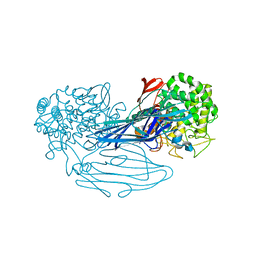 | | The X-ray crystallographic structure of amylo-alpha-1,6-glucosidase from Thermococcus gammatolerans STB12 | | Descriptor: | Amylo-alpha-1,6-glucosidase, putative archaeal type glycogen debranching enzyme (Gde) | | Authors: | Li, Z.F, Ban, X.F, Wang, Y.M, Li, C.M, Gu, Z.B. | | Deposit date: | 2021-10-08 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | The X-ray Crystallographic Structure of Amylo-alpha-1,6-glucosidase from Thermococcus gannatilerans STB12
To Be Published
|
|
7W52
 
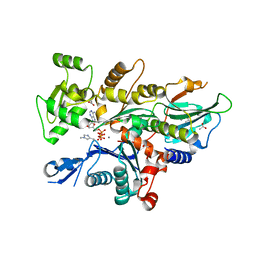 | |
7VLI
 
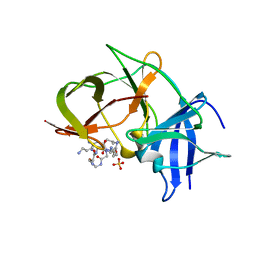 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2220 | | Descriptor: | 1-[(3~{S},6~{S},19~{R})-3,6-bis(4-azanylbutyl)-2,5,8,12,15,18-hexakis(oxidanylidene)-1,4,7,11,14,17-hexazacyclotricos-19-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7VLG
 
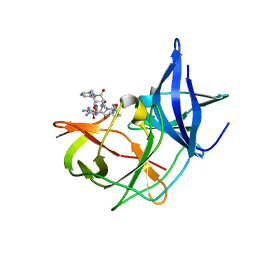 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2201 | | Descriptor: | 1-[(5~{R},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-4,7,14,17,20-pentakis(oxidanylidene)-5-propan-2-yl-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7VLH
 
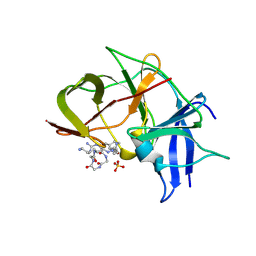 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2219 | | Descriptor: | 1-[(3~{S},6~{R},18~{R})-3,6-bis(4-azanylbutyl)-2,5,8,11,14,17-hexakis(oxidanylidene)-1,4,7,10,13,16-hexazacyclodocos-18-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7VTK
 
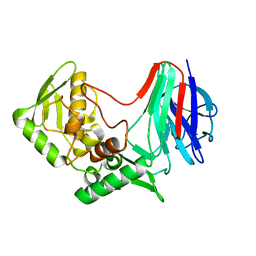 | |
7VUD
 
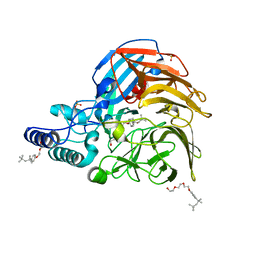 | |
2N1T
 
 | | Dynamic binding mode of a synaptotagmin-1-SNARE complex in solution | | Descriptor: | Synaptosomal-associated protein 25, Synaptotagmin-1, Syntaxin-1A, ... | | Authors: | Brewer, K, Bacaj, T, Cavalli, A, Camilloni, C, Swarbrick, J, Liu, J, Zhou, A, Zhou, P, Barlow, N, Xu, J, Seven, A, Prinslow, E, Voleti, R, Haussinger, D, Bonvin, A, Tomchick, D, Vendruscolo, M, Graham, B, Sudhof, T, Rizo, J. | | Deposit date: | 2015-04-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1TCX
 
 | | HIV TRIPLE MUTANT PROTEASE COMPLEXED WITH INHIBITOR SB203386 | | Descriptor: | (2R,4S,5S,1'S)-2-PHENYLMETHYL-4-HYDROXY-5-(TERT-BUTOXYCARBONYL)AMINO-6-PHENYL HEXANOYL-N-(1'-IMIDAZO-2-YL)-2'-METHYLPROPANAMIDE, HIV PROTEASE | | Authors: | Hoog, S.S, Abdel-Meguid, S.S. | | Deposit date: | 1996-06-05 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human immunodeficiency virus protease ligand specificity conferred by residues outside of the active site cavity.
Biochemistry, 35, 1996
|
|
2N1Q
 
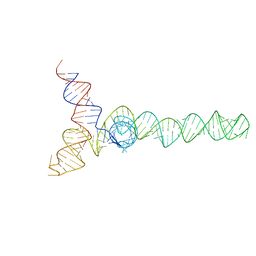 | | HIV-1 Core Packaging Signal | | Descriptor: | RNA_(155-MER) | | Authors: | Keane, S.C, Summers, M.F. | | Deposit date: | 2015-04-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RNA structure. Structure of the HIV-1 RNA packaging signal.
Science, 348, 2015
|
|
2MSX
 
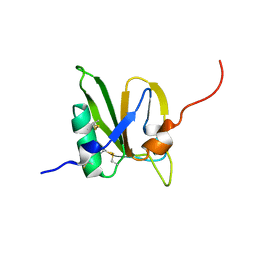 | | The solution structure of the MANEC-type domain from Hepatocyte Growth Factor Inhibitor 1 reveals an unexpected PAN/apple domain-type fold | | Descriptor: | Kunitz-type protease inhibitor 1 | | Authors: | Hong, Z, Nowakowski, M.E, Spronk, C, Petersen, S.V, Petersen, J.S, Kozminski, W, Mulder, F, Jensen, J.K. | | Deposit date: | 2014-08-09 | | Release date: | 2014-12-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the MANEC-type domain from hepatocyte growth factor activator inhibitor-1 reveals an unexpected PAN/apple domain-type fold.
Biochem.J., 466, 2015
|
|
2N8Y
 
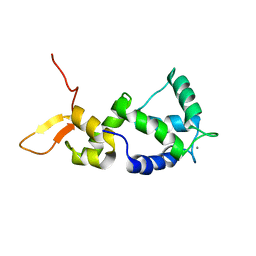 | | Holo form of Calmodulin-Like Domain of Human Non-Muscle alpha-actinin 1 | | Descriptor: | Alpha-actinin-1, CALCIUM ION | | Authors: | Drmota Prebil, S, Slapsak, U, de Almeida Ribeiro, E, Pavsic, M, Ilc, G, Zielinska, K, Hartl, M, Backman, L, Plavec, J, Lenarcic, B, Djinovic-Carugo, K. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and calcium-binding studies of calmodulin-like domain of human non-muscle alpha-actinin-1.
Sci Rep, 6, 2016
|
|
2N4I
 
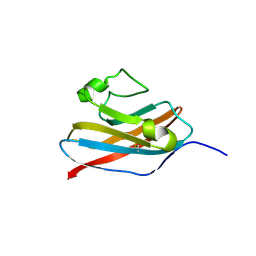 | | The solution structure of Skint-1, a critical determinant of dendritic epidermal gamma-delta T cell selection | | Descriptor: | Selection and upkeep of intraepithelial T-cells protein 1 | | Authors: | Salim, M, Knowles, T.J, Hart, R, Mohammed, F, Woodward, M.J, Willcox, C.R, Overduin, M, Hayday, A.C, Willcox, B.E. | | Deposit date: | 2015-06-18 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Putative Receptor Binding Surface on Skint-1, a Critical Determinant of Dendritic Epidermal T Cell Selection.
J.Biol.Chem., 291, 2016
|
|
4YOL
 
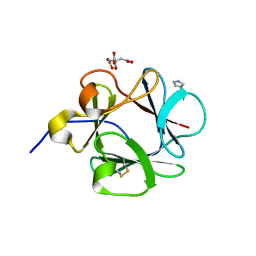 | | Human fibroblast growth factor-1 C16S/A66C/C117A/P134A | | Descriptor: | CITRATE ANION, Fibroblast growth factor 1, IMIDAZOLE | | Authors: | Xia, X, Blaber, M. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Engineering a Cysteine-Free Form of Human Fibroblast Growth Factor-1 for "Second Generation" Therapeutic Application.
J.Pharm.Sci., 105, 2016
|
|
1UC5
 
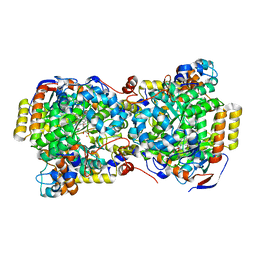 | | Structure of diol dehydratase complexed with (R)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.Biol.Chem., 278, 2003
|
|
7WLG
 
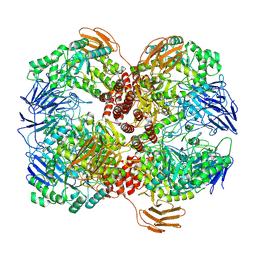 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | Descriptor: | Alpha-xylosidase | | Authors: | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
9CV7
 
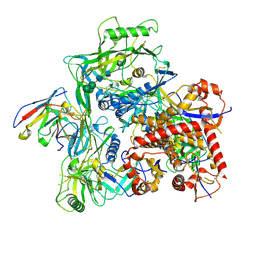 | | LJF-085 Fab in complex with HIV Env ZM233 NFL TD CC3+ trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LJF-085 heavy chain Fv, ... | | Authors: | Ozorowski, G, Ward, A.B. | | Deposit date: | 2024-07-28 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Vaccination of nonhuman primates elicits a broadly neutralizing antibody lineage targeting a quaternary epitope on the HIV-1 Env trimer.
Immunity, 58, 2025
|
|
2NLA
 
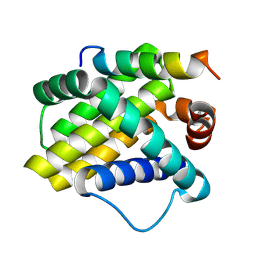 | | Crystal structure of the Mcl-1:mNoxaB BH3 complex | | Descriptor: | FUSION PROTEIN CONSISTING OF Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Phorbol-12-myristate-13-acetate-induced protein 1 | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2006-10-19 | | Release date: | 2007-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2NLE
 
 | | Human beta-defensin-1 (Mutant Gln11Ala) | | Descriptor: | ACETATE ION, Beta-defensin 1, SULFATE ION | | Authors: | Lubkowski, J, Pazgier, M. | | Deposit date: | 2006-10-20 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Studies of the Biological Properties of Human beta-Defensin 1.
J.Biol.Chem., 282, 2007
|
|
9D7G
 
 | | BG505 DS-SOSIP.664 apo structure from the CH103 KN cryo-EM dataset | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface protein gp120, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 33, 2025
|
|
9D7P
 
 | | Cryo-EM structure of BG505 DS-SOSIP.664 with 2 CH103 Fabs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH103 Fab heavy chain, ... | | Authors: | Parsons, R.J, Acharya, P. | | Deposit date: | 2024-08-16 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Acquisition of quaternary trimer interaction as a key step in the lineage maturation of a broad and potent HIV-1 neutralizing antibody.
Structure, 33, 2025
|
|
