7MQM
 
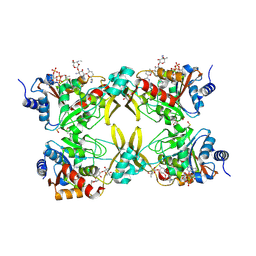 | | AAC(3)-IIIa in complex with CoA and gentamicin | | Descriptor: | Aminoglycoside N(3)-acetyltransferase III, COENZYME A, GLYCEROL, ... | | Authors: | Zielinski, M, Berghuis, A.M. | | Deposit date: | 2021-05-05 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural elucidation of substrate-bound aminoglycoside acetyltransferase (3)-IIIa.
Plos One, 17, 2022
|
|
7MQL
 
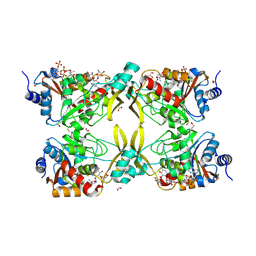 | | AAC(3)-IIIa in complex with CoA and neomycin | | Descriptor: | Aminoglycoside N(3)-acetyltransferase III, COENZYME A, FORMIC ACID, ... | | Authors: | Zielinski, M, Berghuis, A.M. | | Deposit date: | 2021-05-05 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural elucidation of substrate-bound aminoglycoside acetyltransferase (3)-IIIa.
Plos One, 17, 2022
|
|
3Q27
 
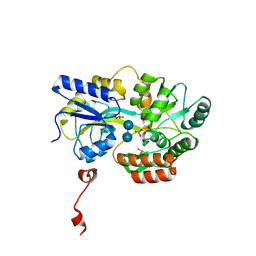 | | Cyrstal structure of human alpha-synuclein (32-57) fused to maltose binding protein (MBP) | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, ... | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
4BBA
 
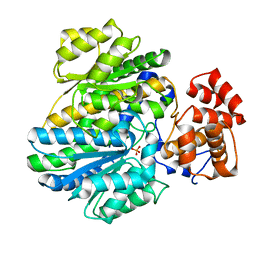 | | Crystal structure of glucokinase regulatory protein complexed to phosphate | | Descriptor: | GLUCOKINASE REGULATORY PROTEIN, PHOSPHATE ION | | Authors: | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinhart, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | Deposit date: | 2012-09-21 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
3Q2X
 
 | |
4BG9
 
 | | Apo form of a putative sugar kinase MK0840 from Methanopyrus kandleri (orthorhombic space group) | | Descriptor: | ACETATE ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Schacherl, M, Waltersperger, S.M, Baumann, U. | | Deposit date: | 2013-03-24 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Characterization of the Ribonuclease H-Like Type Askha Superfamily Kinase Mk0840 from Methanopyrus Kandleri
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BLJ
 
 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 2 | | Descriptor: | Bis-(3-deoxy-3-(3-methoxy-benzamido)-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
1UG3
 
 | | C-terminal portion of human eIF4GI | | Descriptor: | eukaryotic protein synthesis initiation factor 4G | | Authors: | Bellsolell, L, Cho-Park, P.F, Poulin, F, Sonenberg, N, Burley, S.K. | | Deposit date: | 2003-06-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1
Structure, 14, 2006
|
|
1TOW
 
 | | Crystal structure of human adipocyte fatty acid binding protein in complex with a carboxylic acid ligand | | Descriptor: | 4-(9H-CARBAZOL-9-YL)BUTANOIC ACID, Fatty acid-binding protein, adipocyte | | Authors: | Lehmann, F, Haile, S, Axen, E, Medina, C, Uppenberg, J, Svensson, S, Lundback, T, Rondahl, L, Barf, T. | | Deposit date: | 2004-06-15 | | Release date: | 2004-08-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of inhibitors of human adipocyte fatty acid-binding protein, a potential type 2 diabetes target.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1TVZ
 
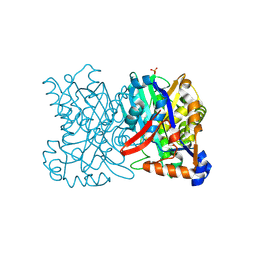 | | Crystal structure of 3-hydroxy-3-methylglutaryl-coenzyme A synthase from Staphylococcus aureus | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, SULFATE ION | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
1TWG
 
 | | RNA polymerase II complexed with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
3ZY1
 
 | |
3ZYH
 
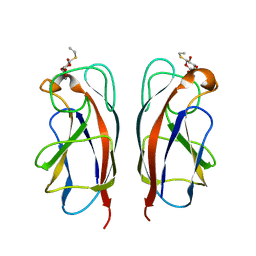 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALBG0 AT 1.5 A RESOLUTION | | Descriptor: | 3-(beta-D-galactopyranosylthio)propanoic acid, CALCIUM ION, PA-I galactophilic lectin | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3ZYB
 
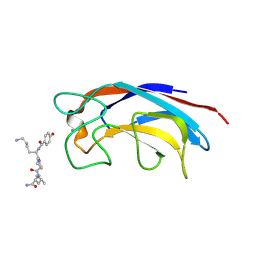 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALAG0 AT 2.3 A RESOLUTION | | Descriptor: | CALCIUM ION, GALA-LYS-PRO-LEUNH2, P-HYDROXYBENZOIC ACID, ... | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-19 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3ZYF
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH NPG AT 1.9 A RESOLUTION | | Descriptor: | 4-nitrophenyl beta-D-galactopyranoside, CALCIUM ION, PA-I GALACTOPHILIC LECTIN | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3NI3
 
 | | 54-Membered ring macrocyclic beta-sheet peptide | | Descriptor: | 54-membered ring macrocyclic beta-sheet peptide, ISOPROPYL ALCOHOL | | Authors: | Sawaya, M.R, Eisenberg, D, Nowick, J.S, Korman, T.P, Khakshoor, O. | | Deposit date: | 2010-06-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray crystallographic structure of an artificial beta-sheet dimer.
J.Am.Chem.Soc., 132, 2010
|
|
1TBP
 
 | |
1TOK
 
 | | Maleic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, MALEIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
3ZC3
 
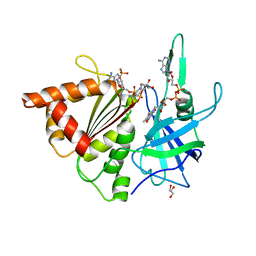 | | FERREDOXIN-NADP REDUCTASE (MUTATION S80A) COMPLEXED WITH NADP BY COCRYSTALLIZATION | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martinez-Julvez, M, Hurtado-Guerrero, R, Herguedas, B, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Hydrogen Bond Network in the Active Site of Anabaena Ferredoxin-Nadp(+) Reductase Modulates its Catalytic Efficiency.
Biochim.Biophys.Acta, 1837, 2013
|
|
1TOJ
 
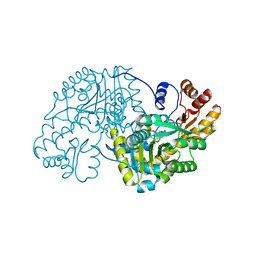 | | Hydrocinnamic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOO
 
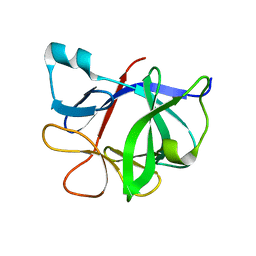 | | Interleukin 1B Mutant F146W | | Descriptor: | Interleukin-1 beta | | Authors: | Adamek, D.H, Guerrero, L, Caspar, D.L. | | Deposit date: | 2004-06-14 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and energetic consequences of mutations in a solvated hydrophobic cavity.
J.Mol.Biol., 346, 2005
|
|
1TP0
 
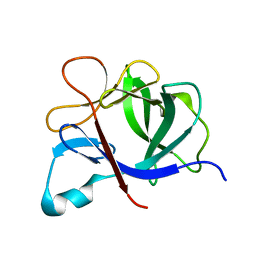 | |
3PPD
 
 | | GGVLVN segment from Human Prostatic Acid Phosphatase Residues 260-265, involved in Semen-Derived Enhancer of Viral Infection | | Descriptor: | ACETIC ACID, GGVLVN peptide, amyloid forming segment, ... | | Authors: | Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation.
Nature, 475, 2011
|
|
1TR7
 
 | | FimH adhesin receptor binding domain from uropathogenic E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, FimH protein, ... | | Authors: | Bouckaert, J, Berglund, J, Schembri, M, De Genst, E, Cools, L, Wuhrer, M, Hung, C.S, Pinkner, J, Slattegard, R, Zavialov, A, Choudhury, D, Langermann, S, Hultgren, S.J, Wyns, L, Klemm, P, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-06-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin
Mol.Microbiol., 55, 2005
|
|
3ZUO
 
 | | OMCI in complex with leukotriene B4 | | Descriptor: | COMPLEMENT INHIBITOR, LEUKOTRIENE B4 | | Authors: | Roversi, P, Maillet, I, Togbe, D, Couillin, I, Quesniaux, V.F.J, Teixeira, M, Ahmat, N, Lissina, O, Boland, W, Ploss, K, Caesar, J.J.E, Leonhartsberger, S, Ryffel, B, Lea, S.M, Nunn, M.A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Bifunctional Lipocalin Ameliorates Murine Immune Complex-Induced Acute Lung Injury.
J.Biol.Chem., 288, 2013
|
|
