6TCC
 
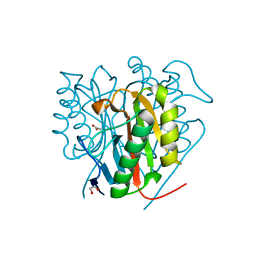 | | Crystal structure of Salmo salar RidA-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ricagno, S, Visentin, C, Di Pisa, F, Digiovanni, S, Oberti, L, Degani, G, Popolo, L, Bartorelli, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage.
Sci Rep, 10, 2020
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
1KX5
 
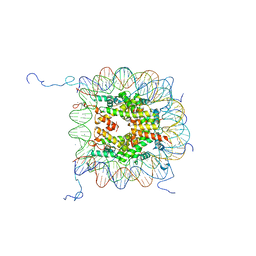 | | X-Ray Structure of the Nucleosome Core Particle, NCP147, at 1.9 A Resolution | | Descriptor: | CHLORIDE ION, DNA (5'(ATCAATATCCACCTGCAGATACTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGGAATCCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGTATCTGCAGGTGGATATTGAT)3'), DNA (5'(ATCAATATCCACCTGCAGATACTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCAGCTGGATTCCAGCTGAACATGCCTTTTGATGGAGCAGTTTCCAAATACACTTTTGGTAGTATCTGCAGGTGGATATTGAT)3'), ... | | Authors: | Davey, C.A, Sargent, D.F, Luger, K, Maeder, A.W, Richmond, T.J. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 A Resolution
J.Mol.Biol., 319, 2002
|
|
9CMQ
 
 | |
9CQK
 
 | |
6E04
 
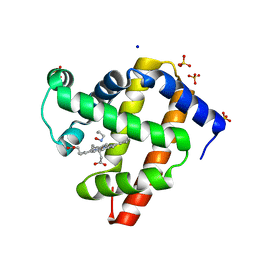 | | sperm whale myoglobin 1-nitrosopropane | | Descriptor: | 1-nitrosopropane, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herrera, V.E. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Nitrosoalkane Binding to Myoglobin Provided by Crystallography of Wild-Type and Distal Pocket Mutant Derivatives.
Biochemistry, 62, 2023
|
|
6NU8
 
 | |
4E4U
 
 | | Crystal structure of a putative Mandelate racemase/Muconate lactonizing enzyme (Target PSI-200780) from Burkholderia SAR-1 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Mandalate racemase/muconate lactonizing enzyme | | Authors: | Kumar, P.R, Bonanno, J, Chowdhury, S, Foti, R, Gizzi, A, Hammonds, J, Hillerich, B, Matikainen, B, Seidel, R, Toro, R, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-13 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a putative MR/ML enzyme from Burkholderia SAR-1
to be published
|
|
4XYK
 
 | | Crystal structure of human phosphofructokinase-1 in complex with ADP, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
9CKN
 
 | | Histidine-covalent alpha-helical peptide (compound 6) targeting hMcl-1 | | Descriptor: | Helical Peptide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Muzzarelli, K.M, Assar, Z, Alboreggia, G, Pellecchia, M. | | Deposit date: | 2024-07-09 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Covalent Targeting of Histidine Residues with Aryl Fluorosulfates: Application to Mcl-1 BH3 Mimetics.
J.Med.Chem., 67, 2024
|
|
3TSZ
 
 | |
7O2V
 
 | | AURORA KINASE A IN COMPLEX WITH THE AUR-A/PDK1 INHIBITOR VI8 | | Descriptor: | 1-[[3,4-bis(fluoranyl)phenyl]methyl]-~{N}-[(1~{R})-2-[[(3~{E})-3-(1~{H}-imidazol-5-ylmethylidene)-2-oxidanylidene-1~{H}-indol-5-yl]amino]-2-oxidanylidene-1-phenyl-ethyl]-6-methyl-2-oxidanylidene-pyridine-3-carboxamide, Aurora kinase A | | Authors: | Garau, G. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of potent dual PDK1/AurA kinase inhibitors for cancer therapy: Lead-optimization, structural insights, and ADME-Tox profile.
Eur.J.Med.Chem., 226, 2021
|
|
1SZU
 
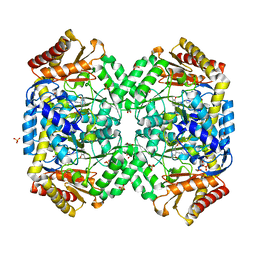 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
2NCS
 
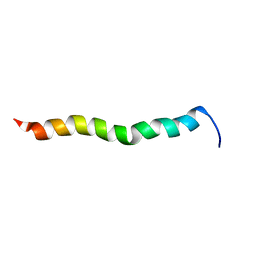 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2YJQ
 
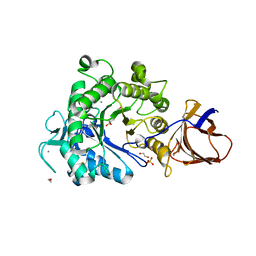 | | Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CEL44C, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-05-23 | | Release date: | 2011-06-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3U6Z
 
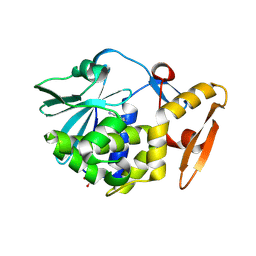 | | Crystal structure of the complex formed between type 1 ribosome inactivating protein and adenine at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states
Biochim.Biophys.Acta, 1824, 2012
|
|
6ZZY
 
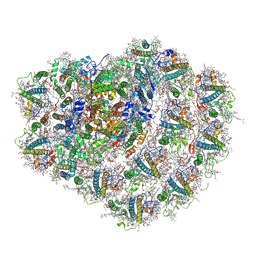 | | Structure of high-light grown Chlorella ohadii photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
5FUU
 
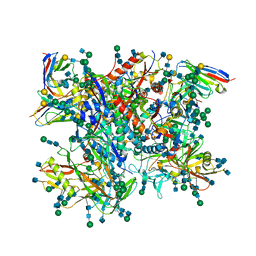 | | Ectodomain of cleaved wild type JR-FL EnvdCT trimer in complex with PGT151 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, J.H, Ward, A.B. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Cryo-Em Structure of a Native, Fully Glycosylated and Cleaved HIV-1 Envelope Trimer
Science, 351, 2016
|
|
7O4G
 
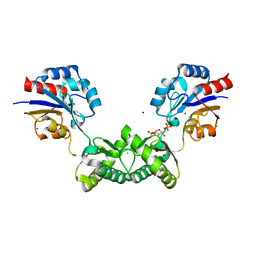 | | Human phosphomannomutase 2 (PMM2) wild-type soaked with the activator glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ramon-Maiques, S, Briso-Montiano, A, Del Cano-Ochoa, F, Vilas, A, Perez, B, Rubio, V. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insight on molecular pathogenesis and pharmacochaperoning potential in phosphomannomutase 2 deficiency, provided by novel human phosphomannomutase 2 structures.
J Inherit Metab Dis, 45, 2022
|
|
1KQY
 
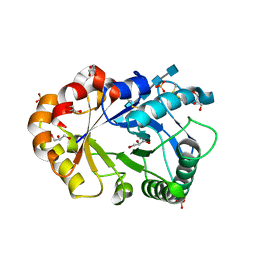 | | Hevamine Mutant D125A/E127A/Y183F in Complex with Penta-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hevamine A, ... | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
7O58
 
 | | Human phosphomannomutase 2 (PMM2) with mutation T237M in complex with the activator glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ramon-Maiques, S, Briso-Montiano, A, Del Cano-Ochoa, F, Vilas, A, Perez, B, Rubio, V. | | Deposit date: | 2021-04-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Insight on molecular pathogenesis and pharmacochaperoning potential in phosphomannomutase 2 deficiency, provided by novel human phosphomannomutase 2 structures.
J Inherit Metab Dis, 45, 2022
|
|
6DHN
 
 | | Bovine glutamate dehydrogenase complexed with Eu3+ | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A novel mechanism of V-type zinc inhibition of glutamate dehydrogenase results from disruption of subunit interactions necessary for efficient catalysis.
FEBS J., 278, 2011
|
|
6DIO
 
 | | Structure of class II HMG-CoA reductase from Delftia acidovorans with NAD bound | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-3-methylglutaryl coenzyme A reductase, CITRIC ACID, ... | | Authors: | Ragwan, E.R, Arai, E, Kung, Y. | | Deposit date: | 2018-05-23 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | New Crystallographic Snapshots of Large Domain Movements in Bacterial 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase.
Biochemistry, 57, 2018
|
|
7OPZ
 
 | | Camel GSTM1-1 in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione transferase, SODIUM ION | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Functional Characterization of Camelus dromedarius Glutathione Transferase M1-1.
Life, 12, 2022
|
|
