3TWF
 
 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3a | | Descriptor: | ACETYL GROUP, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6O3A
 
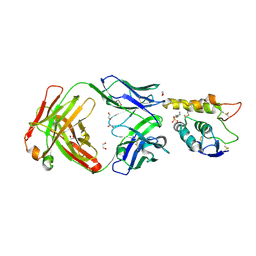 | | Crystal structure of Frizzled 7 CRD in complex with F7.B Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | Authors: | Raman, S, Beilschmidt, M, Fransson, J, Julien, J.P. | | Deposit date: | 2019-02-26 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided design fine-tunes pharmacokinetics, tolerability, and antitumor profile of multispecific frizzled antibodies.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7P18
 
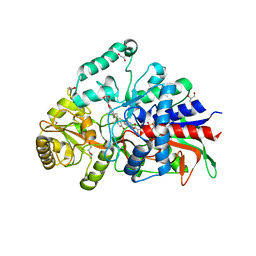 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Sterolibacterium denitrificans in complex with 1,4-androstadiene-3,17-dione | | Descriptor: | 3-oxosteroid 1-dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wojcik, P, Mrugala, B, Kurpiewska, K, Szaleniec, M. | | Deposit date: | 2021-07-01 | | Release date: | 2021-07-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure, Mutagenesis, and QM:MM Modeling of 3-Ketosteroid Delta 1 -Dehydrogenase from Sterolibacterium denitrificans ─The Role of a New Putative Membrane-Associated Domain and Proton-Relay System in Catalysis.
Biochemistry, 62, 2023
|
|
6NYQ
 
 | |
7P2H
 
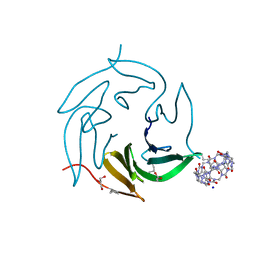 | | Dimethylated fusion protein of RSL and mussel adhesion peptide (Mefp) in complex with cucurbit[7]uril, H3 sheet assembly | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SODIUM ION, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Segregated Protein-Cucurbit[7]uril Crystalline Architectures via Modulatory Peptide Tectons.
Chemistry, 27, 2021
|
|
7P2J
 
 | | Dimethylated fusion protein of RSL and trimeric coiled coil (4dzn) in complex with cucurbit[7]uril, H3 sheet assembly | | Descriptor: | Fucose-binding lectin protein, SODIUM ION, beta-D-mannopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Segregated Protein-Cucurbit[7]uril Crystalline Architectures via Modulatory Peptide Tectons.
Chemistry, 27, 2021
|
|
6O3C
 
 | | Crystal structure of active Smoothened bound to SAG21k, cholesterol, and NbSmo8 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-4,7-difluoro-N-{[2-methoxy-5-(pyridin-4-yl)phenyl]methyl}-N-[trans-4-(methylamino)cyclohexyl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Deshpande, I.S, Liang, J, Hedeen, D, Roberts, K.J, Zhang, Y, Ha, B, Latorraca, N.R, Faust, B, Dror, R.O, Beachy, P.A, Myers, B.R, Manglik, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Smoothened stimulation by membrane sterols drives Hedgehog pathway activity.
Nature, 571, 2019
|
|
7OKW
 
 | | 1.62A X-ray crystal structure of the conserved C-terminal (CCT) of human OSR1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bax, B.D, Elvers, K.T, Lipka-Lloyd, M, Mehellou, Y. | | Deposit date: | 2021-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of the Human SPAK and OSR1 Conserved C-Terminal (CCT) Domains.
Chembiochem, 23, 2022
|
|
7P51
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2 MAIN PROTEASE COMPLEXED WITH FRAGMENT F01 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-chloropyridin-2-yl)-3-oxo-2,3-dihydro-1H-indene-1-carboxamide, ... | | Authors: | Hanoulle, X, Moschidi, D. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OKC
 
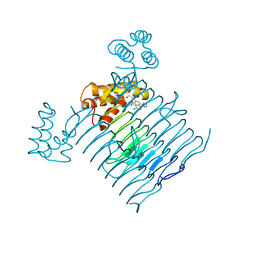 | | Crystal structure of Escherichia coli LpxA in complex with compound 1 | | Descriptor: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, SODIUM ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7P1S
 
 | | Structure of KDNase from Trichophyton Rubrum in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, Extracellular sialidase/neuraminidase, SODIUM ION | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1R
 
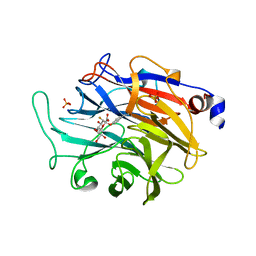 | |
7P8Z
 
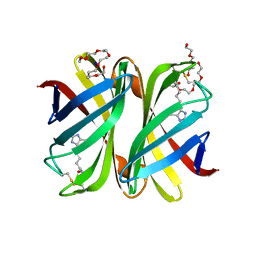 | | Short wilavidin biotin complex | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, BIOTIN, ... | | Authors: | Avraham, O, Livnah, O. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Wilavidin - a novel member of the avidin family that forms unique biotin-binding hexamers.
Febs J., 289, 2022
|
|
6LDW
 
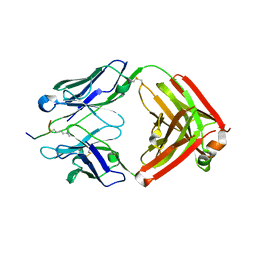 | |
7OV3
 
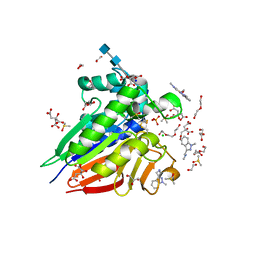 | | Crystal structure of pig purple acid phosphatase in complex with Maybridge fragment CC063346, dimethyl sulfoxide and citrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
7P0D
 
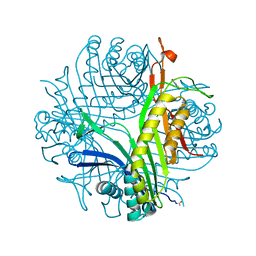 | |
7P0G
 
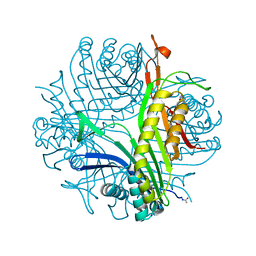 | |
7P0C
 
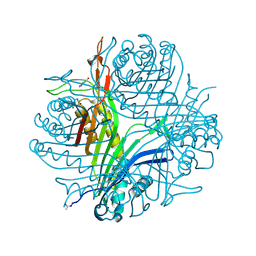 | |
7OPZ
 
 | | Camel GSTM1-1 in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione transferase, SODIUM ION | | Authors: | Papageorgiou, A.C, Poudel, N. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Functional Characterization of Camelus dromedarius Glutathione Transferase M1-1.
Life, 12, 2022
|
|
7OPY
 
 | |
7P9Q
 
 | | Crystal structure of Indole 3-Carboxylic acid decarboxylase from Arthrobacter nicotianae FI1612 in complex with co-factor prFMN. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, AnInD, MANGANESE (II) ION, ... | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2021-07-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and biochemical characterization of the prenylated flavin mononucleotide-dependent indole-3-carboxylic acid decarboxylase.
J.Biol.Chem., 298, 2022
|
|
7P3I
 
 | | Crystal structure of human CD40/TNFRSF5 in complex with the anti-CD40 DARPin protein | | Descriptor: | Darpin, SODIUM ION, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Malvezzi, F, Mangold, S, Hospodarsch, T, Reichen, C, Iss, C, Lammens, A, Krapp, S, Domke, C. | | Deposit date: | 2021-07-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Multispecific Anti-CD40 DARPin Construct Induces Tumor-Selective CD40 Activation and Tumor Regression.
Cancer Immunol Res, 10, 2022
|
|
7OV8
 
 | | Crystal structure of pig purple acid phosphatase in complex with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and glycerol | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
7P0X
 
 | | Crystal structure of Thioredoxin reductase from Brugia Malayi | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Ardini, M, Silvestri, I, Gabriele, F, Cheng, Q, Arner, E.S.J, Williams, D.L. | | Deposit date: | 2021-06-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Biochemical and structural characterizations of thioredoxin reductase selenoproteins of the parasitic filarial nematodes Brugia malayi and Onchocerca volvulus.
Redox Biol, 51, 2022
|
|
6O6A
 
 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, ligand-free state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, SODIUM ION, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Diver, M.M, Cheng, Y, Julius, D. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
