5UL5
 
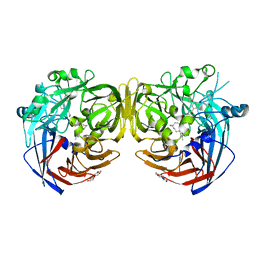 | | Crystal structure of RPE65 in complex with MB-004 and palmitate | | Descriptor: | (1R)-3-amino-1-{3-[(2-propylpentyl)oxy]phenyl}propan-1-ol, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, FE (II) ION, ... | | Authors: | Kiser, P.D, Palczewski, K. | | Deposit date: | 2017-01-24 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Tuning of Visual Cycle Modulator Pharmacodynamics.
J. Pharmacol. Exp. Ther., 362, 2017
|
|
8B6F
 
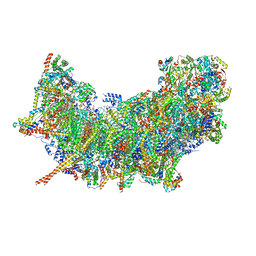 | | Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
6QL9
 
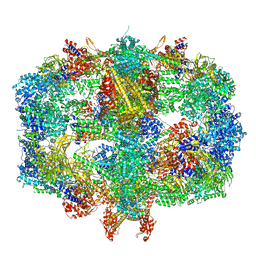 | | Structure of Fatty acid synthase complex from Saccharomyces cerevisiae at 2.9 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, 4'-PHOSPHOPANTETHEINE, ACETATE ION, ... | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
5D86
 
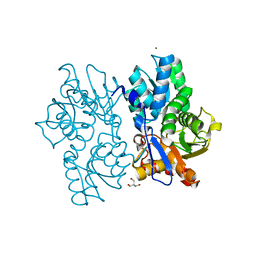 | | Staphyloferrin B precursor biosynthetic enzyme SbnA Y152F variant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kobylarz, M.J, Grigg, J.C, Liu, Y, Lee, M.S.F, Heinrichs, D.E, Murphy, M.E.P. | | Deposit date: | 2015-08-15 | | Release date: | 2016-02-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.
Biochemistry, 55, 2016
|
|
6NZH
 
 | |
5EDN
 
 | | Structure of HOXB13-DNA(TCG) complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(P*GP*GP*AP*CP*CP*TP*CP*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*GP*TP*CP*C)-3'), ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
5H29
 
 | | Crystal Structure of the NTD_N/C domain of Alkylhydroperoxide Reductase AhpF from Enterococcus Faecalis (V583) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PROLINE, SULFATE ION, ... | | Authors: | Balakrishna, A.M, Kwang, T.Y, Gruber, G. | | Deposit date: | 2016-10-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel insights into the vancomycin-resistant Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit F
Biochim. Biophys. Acta, 1861, 2017
|
|
4S18
 
 | |
5UPE
 
 | |
6NZP
 
 | |
5DVY
 
 | | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Penicillin binding protein 2 prime, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-21 | | Release date: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
5M41
 
 | | Crystal structure of nigritoxine | | Descriptor: | MAGNESIUM ION, Nigritoxine | | Authors: | Czjzek, M, Labreuche, L, Jeudy, A, Le Roux, F. | | Deposit date: | 2016-10-17 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nigritoxin is a bacterial toxin for crustaceans and insects.
Nat Commun, 8, 2017
|
|
8U6P
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-((2-cyanoindolizin-8-yl)oxy)phenoxy)-N,N-dimethylpropanamide (JLJ754), a non-nucleoside inhibitor | | Descriptor: | 3-(2-{[(4S)-2-cyanoindolizin-8-yl]oxy}phenoxy)-N,N-dimethylpropanamide, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6N
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-((6-cyanonaphthalen-1-yl)oxy)phenoxy)-N,N-dimethylpropanamide (JLJ752), a non-nucleoside inhibitor | | Descriptor: | 3-{2-[(6-cyanonaphthalen-1-yl)oxy]phenoxy}-N,N-dimethylpropanamide, Gag-Pol polyprotein, p51 RT | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
5LAQ
 
 | | Crystal structure of human phosphodiesterase 4B catalytic domain with inhibitor NPD-001 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-06-14 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
5LBO
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-001 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-06-16 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
2GO0
 
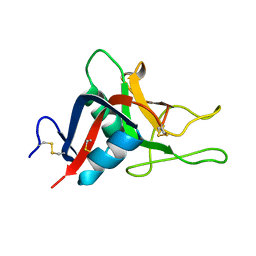 | |
5VFY
 
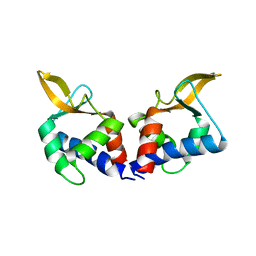 | | Structure of an accessory protein of the pCW3 relaxosome | | Descriptor: | TcpK | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
5DNF
 
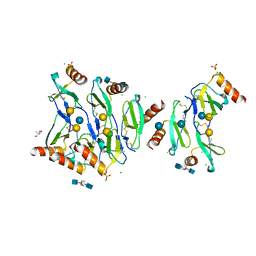 | | Crystal structure of CC chemokine 5 (CCL5) oligomer in complex with heparin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, C-C motif chemokine 5, ... | | Authors: | Liang, W.G, Tang, W. | | Deposit date: | 2015-09-10 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5VFX
 
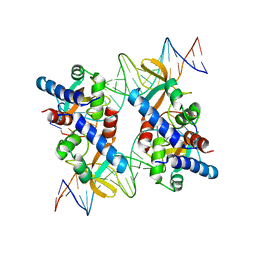 | | Structure of an accessory protein of the pCW3 relaxosome in complex with the origin of transfer (oriT) DNA | | Descriptor: | TcpK, oriT | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
4UFF
 
 | | Thrombin in complex with (2R)-2-(benzylsulfonylamino)-N-(2-((4- carbamimidoylphenyl)methylamino)-2-oxo-ethyl)-N-methyl-3-phenyl- propanamide | | Descriptor: | (2R)-2-(benzylsulfonylamino)-N-(2-((4-carbamimidoylphenyl)methylamino)-2-oxo-ethyl)-N-methyl-3-phenyl-propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-2, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-03-16 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Boosting Affinity by Correct Ligand Preorganization for the S2 Pocket of Thrombin: A Study by Isothermal Titration Calorimetry, Molecular Dynamics, and High-Resolution Crystal Structures.
Chemmedchem, 11, 2016
|
|
4UFG
 
 | | Thrombin in complex with (2R)-2-(benzylsulfonylamino)-N-((1S)-2-((4- carbamimidoylphenyl)methylamino)-1-methyl-2-oxo-ethyl)-N-methyl-3- phenyl-propanamide ethane | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-2-[methyl-[(2R)-3-phenyl-2-[(phenylmethyl)sulfonylamino]propanoyl]amino]butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-2, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-03-17 | | Release date: | 2016-01-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Boosting Affinity by Correct Ligand Preorganization for the S2 Pocket of Thrombin: A Study by Isothermal Titration Calorimetry, Molecular Dynamics, and High-Resolution Crystal Structures.
Chemmedchem, 11, 2016
|
|
5HOF
 
 | | Crystal structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinopentaose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Lansky, S, Shwartstien, O, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinopentaose
To Be Published
|
|
4NVM
 
 | |
4NIR
 
 | |
