2OF6
 
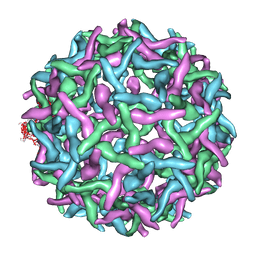 | | Structure of immature West Nile virus | | Descriptor: | envelope glycoprotein E | | Authors: | Zhang, Y, Kaufmann, B, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-01-02 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Structure of immature west nile virus.
J.Virol., 81, 2007
|
|
6HOB
 
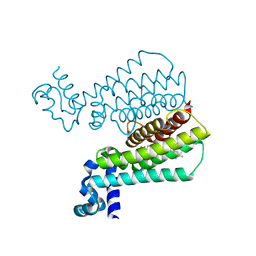 | |
6HO9
 
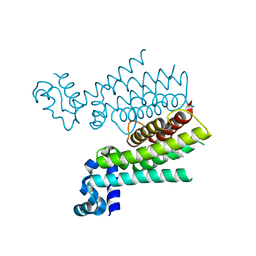 | | TRANSCRIPTIONAL REPRESSOR ETHR FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH BDM44825 | | Descriptor: | 4-[4-(2-methylpropyl)-1,2,3-triazol-1-yl]-~{N}-[3,3,3-tris(fluoranyl)propyl]benzamide, HTH-type transcriptional regulator EthR | | Authors: | Wintjens, R, Wohlkonig, A. | | Deposit date: | 2018-09-17 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A comprehensive analysis of the protein-ligand interactions in crystal structures of Mycobacterium tuberculosis EthR.
Biochim Biophys Acta Proteins Proteom, 1867, 2018
|
|
8PK4
 
 | | Cryo EM structure of the type 5A polymorph of alpha-synuclein. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
7RCP
 
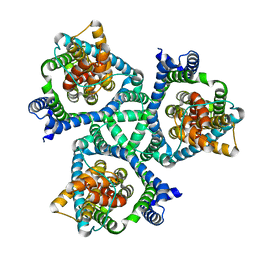 | |
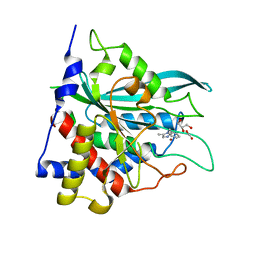
8HY3
 
 | |
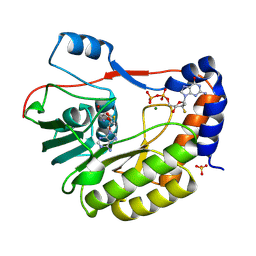
8PEM
 
 | |
6FUL
 
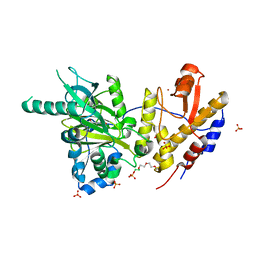 | | Crystal structure of UTX complexed with 5-hydroxy-4-keto-1-methyl-picolinate | | Descriptor: | 1-methyl-5-oxidanyl-4-oxidanylidene-pyridine-2-carboxylic acid, 2-(2-METHOXYETHOXY)ETHANOL, Lysine-specific demethylase 6A, ... | | Authors: | Esposito, C, Sledz, P, Caflisch, A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | In Silico Identification of JMJD3 Demethylase Inhibitors.
J Chem Inf Model, 58, 2018
|
|
6G8F
 
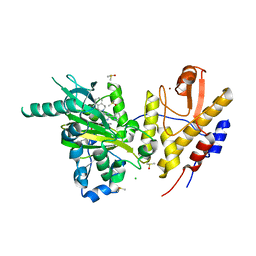 | | Crystal structure of UTX complexed with GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Esposito, C, Sledz, P, Caflisch, A. | | Deposit date: | 2018-04-08 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | In Silico Identification of JMJD3 Demethylase Inhibitors.
J Chem Inf Model, 58, 2018
|
|
7R1U
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R1T
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
6SGH
 
 | |
4MU7
 
 | | Crystal structure of cIAP1 BIR3 bound to T3450325 | | Descriptor: | (3S,10aS)-2-[(2S)-2-cyclohexyl-2-{[(2S)-2-(methylamino)butanoyl]amino}acetyl]-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]-1,2,3,4,10,10a-hexahydropyrazino[1,2-a]indole-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Snell, G.P, Dougan, D.R. | | Deposit date: | 2013-09-20 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design, synthesis, and biological activities of novel hexahydropyrazino[1,2-a]indole derivatives as potent inhibitors of apoptosis (IAP) proteins antagonists with improved membrane permeability across MDR1 expressing cells.
Bioorg.Med.Chem., 21, 2013
|
|
6PZJ
 
 | |
8J26
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-4, AM047-6, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
8J1Q
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-0, AM047-0, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
7R9X
 
 | |
6ROC
 
 | | Crystal structure of Borrelia burgdorferi outer surface protein BBA69, mutant Leu214Met (Se-Met data) | | Descriptor: | Putative surface protein | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2019-05-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Borrelia burgdorferi outer surface protein BBA69 in comparison to the paralogous protein CspA.
Ticks Tick Borne Dis, 10, 2019
|
|
8PQO
 
 | |
2O6P
 
 | | Crystal Structure of the heme-IsdC complex | | Descriptor: | CHLORIDE ION, Iron-regulated surface determinant protein C, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sharp, K.H, Schneider, S, Cockayne, A, Paoli, M. | | Deposit date: | 2006-12-08 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus.
J. Biol. Chem., 282, 2007
|
|
6TNM
 
 | | E. coli aerobic trifunctional enzyme subunit-alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Fatty acid oxidation complex subunit alpha, GLYCEROL, ... | | Authors: | Sah-Teli, S.K, Hynonen, M.J, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2019-12-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Insights into the stability and substrate specificity of the E. coli aerobic beta-oxidation trifunctional enzyme complex.
J.Struct.Biol., 210, 2020
|
|
1KMS
 
 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND 6-([5-QUINOLYLAMINO]METHYL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE (SRI-9439), A LIPOPHILIC ANTIFOLATE | | Descriptor: | 6-([5-QUINOLYLAMINO]METHYL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Klon, A.E, Heroux, A, Ross, L.J, Pathak, V, Johnson, C.A, Piper, J.R, Borhani, D.W. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Atomic structures of human dihydrofolate reductase complexed with NADPH and two lipophilic antifolates at 1.09 a and 1.05 a resolution.
J.Mol.Biol., 320, 2002
|
|
6QDR
 
 | | Crystal structure of 14-3-3sigma in complex with a PAK6 pT99 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Kaplan, A, Fournier, A.E, Ottman, C. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.615 Å) | | Cite: | Polypharmacological Perturbation of the 14-3-3 Adaptor Protein Interactome Stimulates Neurite Outgrowth.
Cell Chem Biol, 27, 2020
|
|
4NJN
 
 | | Crystal Structure of E.coli GlpG at pH 4.5 | | Descriptor: | Rhomboid protease GlpG | | Authors: | Dickey, S.W, Baker, R.P, Cho, S, Urban, S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Proteolysis inside the Membrane Is a Rate-Governed Reaction Not Driven by Substrate Affinity.
Cell(Cambridge,Mass.), 155, 2013
|
|
6V7J
 
 | | The C2221 crystal form of canavalin at 173 K | | Descriptor: | BENZOIC ACID, CALCIUM ION, Canavalin, ... | | Authors: | McPherson, A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of benzoic acid and anions within the cupin domains of the vicilin protein canavalin from jack bean (Canavalia ensiformis): Crystal structures.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
