5MEL
 
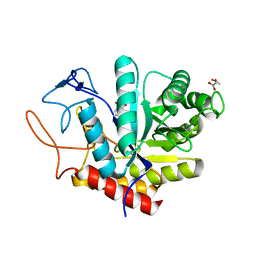 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with Glc-alpha-1,3-(3R,4R,5R)-5-(hydroxymethyl)cyclohex-1,2-ene-3,4-diol | | Descriptor: | (1~{R},2~{R},6~{R})-6-(hydroxymethyl)cyclohex-3-ene-1,2-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5TW9
 
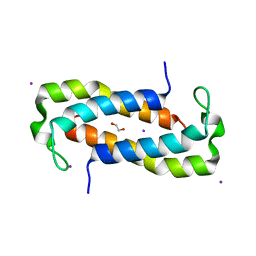 | | 1.50 Angstrom Crystal Structure of C-terminal Fragment (residues 322-384) of Iron Uptake System Component EfeO from Yersinia pestis. | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Iron uptake system component EfeO | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-11 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Crystal Structure of C-terminal Fragment (residues 322-384) of Iron Uptake System Component EfeO from Yersinia pestis.
To Be Published
|
|
8DKG
 
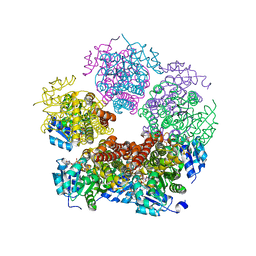 | | Structure of PYCR1 Thr171Met variant complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Isoform 3 of Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Meeks, K.R, Tanner, J.J. | | Deposit date: | 2022-07-05 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional Impact of a Cancer-Related Variant in Human Delta 1 -Pyrroline-5-Carboxylate Reductase 1.
Acs Omega, 8, 2023
|
|
9C6V
 
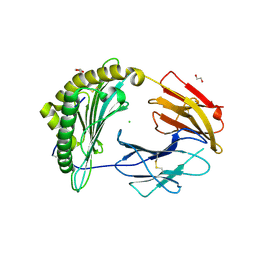 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (1 molecule/asymmetric unit) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
5DU8
 
 | | Crystal structure of M. tuberculosis EchA6 bound to GSK572A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-N-[(5-fluoropyridin-2-yl)methyl]-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5IA9
 
 | | The structure of microsomal glutathione transferase 1 in complex with Meisenheimer complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Microsomal glutathione S-transferase 1, ... | | Authors: | Kuang, Q, Purhonen, P, Jegerschold, C, Morgenstern, R, Hebert, H. | | Deposit date: | 2016-02-21 | | Release date: | 2017-07-12 | | Last modified: | 2025-05-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Dead-end complex, lipid interactions and catalytic mechanism of microsomal glutathione transferase 1, an electron crystallography and mutagenesis investigation.
Sci Rep, 7, 2017
|
|
6ZBW
 
 | | Structure of the D125N mutant of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6-alpha-manno-cyclophellitol trisaccharide inhibitor | | Descriptor: | (1~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, Alpha-1,6-mannanase, ... | | Authors: | Davies, G.J, Offen, W.A. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
6D0P
 
 | | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii
To be Published
|
|
8BOE
 
 | |
8BJX
 
 | |
5L4N
 
 | | Leishmania major Pteridine reductase 1 (PTR1) in complex with compound 1 | | Descriptor: | (2~{R})-2-(3-hydroxyphenyl)-6-oxidanyl-2,3-dihydrochromen-4-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Dello Iacono, L, Di Pisa, F, Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2016-05-26 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
6MK9
 
 | | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Das, D, Mitsuya, H. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-02 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-121
To Be Published
|
|
6P9N
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HT CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH (S)-MCG-IV-210. | | Descriptor: | (3S)-N~1~-(2-aminoethyl)-N~3~-(4-chloro-3-fluorophenyl)piperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-06-10 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
4XZN
 
 | | Crystal structure of the methylated K125R/V301L AKR1B10 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
3DWF
 
 | | Crystal Structure of the Guinea Pig 11beta-Hydroxysteroid Dehydrogenase Type 1 Mutant F278E | | Descriptor: | 11-beta-hydroxysteroid dehydrogenase 1, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lawson, A.J, Ride, J.P, White, S.A, Walker, E.A. | | Deposit date: | 2008-07-22 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutations of key hydrophobic surface residues of 11 beta-hydroxysteroid dehydrogenase type 1 increase solubility and monodispersity in a bacterial expression system
Protein Sci., 18, 2009
|
|
4XZM
 
 | | Crystal structure of the methylated wild-type AKR1B10 holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
6Y3C
 
 | | Human COX-1 Crystal Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Prostaglandin G/H synthase 1, ... | | Authors: | Miciaccia, M, Belviso, B.D, Iaselli, M, Ferorelli, S, Perrone, M.G, Caliandro, R, Scilimati, A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.361 Å) | | Cite: | Three-dimensional structure of human cyclooxygenase (hCOX)-1.
Sci Rep, 11, 2021
|
|
6HMH
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) and alpha-1,2-mannobiose | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Lu, D, Zhu, S, Bernardo-Seisdedos, G, Millet, O, Zhang, Y, Sollogoub, M, Jimenez-Barbero, J, Davies, G.J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | From 1,4-Disaccharide to 1,3-Glycosyl Carbasugar: Synthesis of a Bespoke Inhibitor of Family GH99 Endo-alpha-mannosidase.
Org.Lett., 20, 2018
|
|
4ZN6
 
 | |
6GWR
 
 | | Structure of the kinase domain of human DDR1 in complex with a potent and selective inhibitor of DDR1 and DDR2 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-benzamide, Epithelial discoidin domain-containing receptor 1, ... | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 3-(Imidazo[1,2- a]pyrazin-3-ylethynyl)-4-isopropyl- N-(3-((4-methylpiperazin-1-yl)methyl)-5-(trifluoromethyl)phenyl)benzamide as a Dual Inhibitor of Discoidin Domain Receptors 1 and 2.
J. Med. Chem., 61, 2018
|
|
5WBE
 
 | | COX-1:MOFEZOLAC COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mofezolac, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cingolani, G, Panella, A, Perrone, M.G, Vitale, P, Smith, W.L, Scilimati, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for selective inhibition of Cyclooxygenase-1 (COX-1) by diarylisoxazoles mofezolac and 3-(5-chlorofuran-2-yl)-5-methyl-4-phenylisoxazole (P6).
Eur J Med Chem, 138, 2017
|
|
6S6W
 
 | | Crystal Structure of human ALDH1A3 in complex with 2,6-diphenylimidazo[1,2-a]pyridine (compound GA11) and NAD+ | | Descriptor: | 2,6-diphenylimidazo[1,2-a]pyridine, Aldehyde dehydrogenase family 1 member A3, GLYCEROL, ... | | Authors: | Gelardi, E.L.M, Quattrini, L, LaMotta, C, Ferraris, D.M, Garavaglia, S. | | Deposit date: | 2019-07-03 | | Release date: | 2020-04-22 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Imidazo[1,2-a]pyridine Derivatives as Aldehyde Dehydrogenase Inhibitors: Novel Chemotypes to Target Glioblastoma Stem Cells.
J.Med.Chem., 63, 2020
|
|
5DU6
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK059A. | | Descriptor: | (5R,7R)-5-(4-ethylphenyl)-N-(4-fluorobenzyl)-7-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
4XZL
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0049 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
6SZP
 
 | | High resolution crystal structure of human DDAH-1 in complex with N-(4-Aminobutyl)-N'-(2-Methoxyethyl)guanidine | | Descriptor: | (1~{S})-~{N}'-(4-azanylbutyl)-~{N}"-(2-methoxyethyl)methanetriamine, GLYCEROL, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Hennig, S, Vetter, I.R, Schade, D. | | Deposit date: | 2019-10-02 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery ofN-(4-Aminobutyl)-N'-(2-methoxyethyl)guanidine as the First Selective, Nonamino Acid, Catalytic Site Inhibitor of Human Dimethylarginine Dimethylaminohydrolase-1 (hDDAH-1).
J.Med.Chem., 63, 2020
|
|
