5OCE
 
 | | THE MOLECULAR MECHANISM OF SUBSTRATE RECOGNITION AND CATALYSIS OF THE MEMBRANE ACYLTRANSFERASE PatA -- Complex of PatA with palmitate, mannose, and palmitoyl-6-mannose | | Descriptor: | PALMITIC ACID, Phosphatidylinositol mannoside acyltransferase, [(2~{R},3~{S},4~{S},5~{S},6~{S})-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hexadecanoate, ... | | Authors: | Albesa-Jove, D, Tersa, M, Guerin, M.E. | | Deposit date: | 2017-06-30 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Molecular Mechanism of Substrate Recognition and Catalysis of the Membrane Acyltransferase PatA from Mycobacteria.
ACS Chem. Biol., 13, 2018
|
|
3E3U
 
 | | Crystal structure of Mycobacterium tuberculosis peptide deformylase in complex with inhibitor | | Descriptor: | N-[(2R)-2-{[(2S)-2-(1,3-benzoxazol-2-yl)pyrrolidin-1-yl]carbonyl}hexyl]-N-hydroxyformamide, NICKEL (II) ION, Peptide deformylase | | Authors: | Meng, W, Xu, M, Pan, S, Koehn, J. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3E3A
 
 | | The Structure of Rv0554 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, POSSIBLE PEROXIDASE BPOC | | Authors: | Johnston, J.M, Baker, E.N. | | Deposit date: | 2008-08-06 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of Rv0554 from Mycobacterium tuberculosis: testing a putative role in menaquinone biosynthesis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1E0W
 
 | | Xylanase 10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution | | Descriptor: | ENDO-1,4-BETA-XYLANASE A | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
3FGP
 
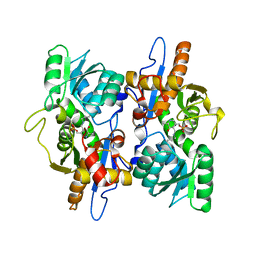 | |
3FIG
 
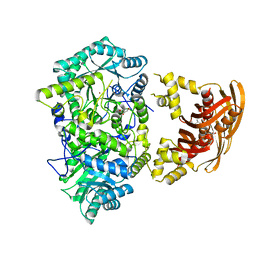 | | Crystal Structure of Leucine-bound LeuA from Mycobacterium tuberculosis | | Descriptor: | 2-isopropylmalate synthase, GLYCEROL, LEUCINE, ... | | Authors: | Koon, N, Squire, C.J, Baker, E.N. | | Deposit date: | 2008-12-11 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of LeuA from Mycobacterium tuberculosis, a key enzyme in leucine biosynthesis.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4GUZ
 
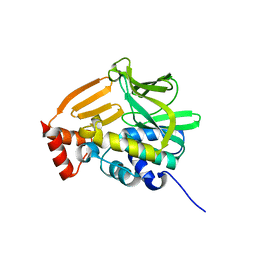 | | Structure of the arylamine N-acetyltransferase from Mycobacterium abscessus | | Descriptor: | Probable arylamine n-acetyl transferase | | Authors: | Kubiak, X, Li de la Sierra-Gallay, I, Haouz, A, Weber, P, Rodrigues-Lima, F. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional characterization of an arylamine N-acetyltransferase from the pathogen Mycobacterium abscessus: differences from other mycobacterial isoforms and implications for selective inhibition.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6EDW
 
 | |
6EE1
 
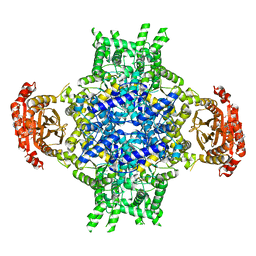 | |
2IDZ
 
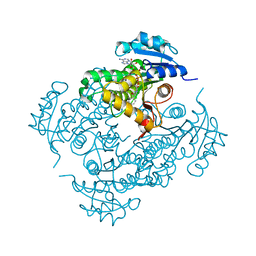 | | Crystal structure of wild type Enoyl-ACP(CoA) reductase from Mycobacterium tuberculosis in complex with NADH-INH | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], ISONICOTINIC-ACETYL-NICOTINAMIDE-ADENINE DINUCLEOTIDE | | Authors: | Dias, M.V.B, Prado, M.P.X, Vasconcelos, I.B, Valmir, F, Basso, L.A, Santos, D.S, Azevedo Jr, W.F. | | Deposit date: | 2006-09-15 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis.
J.Struct.Biol., 159, 2007
|
|
2IE0
 
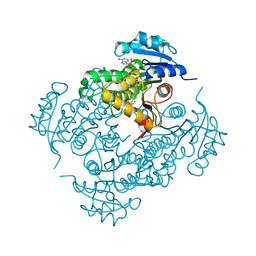 | | Crystal Structure of Isoniazid-resistant I21V Enoyl-ACP(COA) Reductase Mutant Enzyme From MYCOBACTERIUM TUBERCULOSIS in Complex with NADH-INH | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], ISONICOTINIC-ACETYL-NICOTINAMIDE-ADENINE DINUCLEOTIDE | | Authors: | Dias, M.V.B, Prado, A.M.X, Vasconceles, I.B, Fadel, F, Basso, L.A, Santos, D.S, Azevedo Jr, W.F. | | Deposit date: | 2006-09-15 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis.
J.Struct.Biol., 159, 2007
|
|
2IEB
 
 | | Crystal Structure of Isoniazid-resistant S94A ENOYL-ACP(COA) Reductase Mutant Enzyme from MYCOBACTERIUM TUBERCULOSIS in Complex with NADH-INH | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], ISONICOTINIC-ACETYL-NICOTINAMIDE-ADENINE DINUCLEOTIDE | | Authors: | Dias, M.V.B, Prado, A.M.X, Vasconcelos, I.B, Fadel, V, Basso, L.A, Santos, D.S, Azevedo Jr, W.F. | | Deposit date: | 2006-09-18 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis.
J.Struct.Biol., 159, 2007
|
|
2IED
 
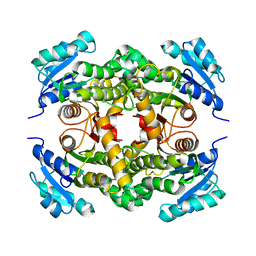 | | CRYSTAL STRUCTURE of ISONIAZID-RESISTANT S94A ENOYL-ACP(COA) REDUCTASE MUTANT ENZYME FROM MYCOBACTERIUM TUBERCULOSIS UNCOMPLEXED | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Dias, M.V.B, Prado, A.M.X, Vasconcelos, I.B, Fadel, V, Basso, L.A, Santos, D.S, Azevedo Jr, W.F. | | Deposit date: | 2006-09-18 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis.
J.Struct.Biol., 159, 2007
|
|
7YST
 
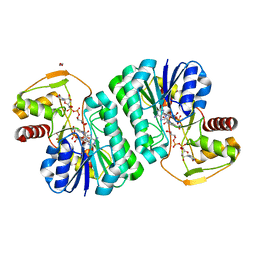 | |
3LAP
 
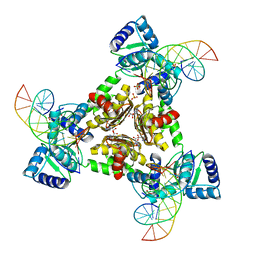 | | The Structure of the Intermediate Complex of the Arginine Repressor from Mycobacterium tuberculosis Bound to its DNA Operator and L-canavanine. | | Descriptor: | 5'-D(*TP*TP*GP*CP*AP*TP*AP*AP*CP*GP*AP*TP*GP*CP*AP*A)-3', 5'-D(*TP*TP*GP*CP*AP*TP*CP*GP*TP*TP*AP*TP*GP*CP*AP*A)-3', Arginine repressor, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-01-06 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | crystal structure of the intermediate complex of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator reveals detailed mechanism of arginine repression.
J.Mol.Biol., 399, 2010
|
|
7OY9
 
 | |
5LIE
 
 | |
5LI7
 
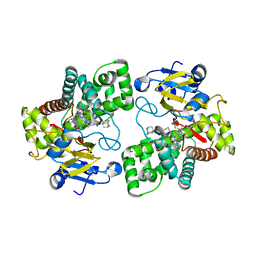 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with 1-(3-(1H-imidazol-1-yl)propyl)-3-((3s,5s,7s)-adamantan-1-yl)urea | | Descriptor: | 1-(1-adamantyl)-3-(3-imidazol-1-ylpropyl)urea, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126 | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
7LHL
 
 | |
5LI8
 
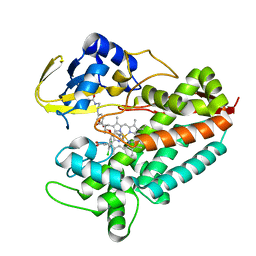 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with ketoconazole | | Descriptor: | 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126 | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
3MAY
 
 | |
5LI6
 
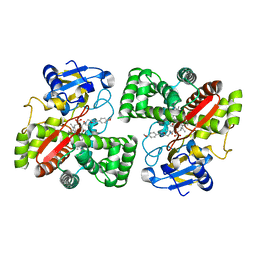 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with N-isopropyl-N-((3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl)methyl)-2-(4-nitrophenyl)acetamide | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126, ~{N}-[[3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl]methyl]-2-(4-nitrophenyl)-~{N}-propan-2-yl-ethanamide | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
5CRF
 
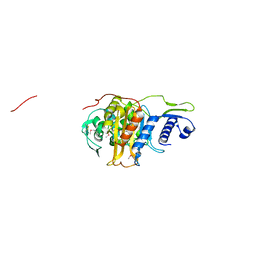 | | Structure of the penicillin-binding protein PonA1 from Mycobacterium Tuberculosis | | Descriptor: | PHOSPHATE ION, Penicillin-binding protein 1A | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-22 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
5D19
 
 | |
5D18
 
 | | Crystal structure of Mycobacterium tuberculosis Rv0302, form I | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis transcriptional regulator Rv0302.
Protein Sci., 2015
|
|
