1KSP
 
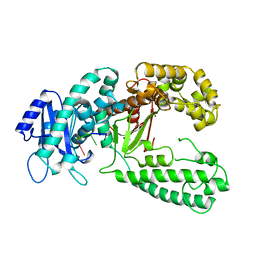 | | DNA polymerase I Klenow fragment (E.C.2.7.7.7) mutant/DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*PST)-3'), PROTEIN (DNA POLYMERASE I-KLENOW FRAGMENT (E.C.2.7.7.7)), ZINC ION | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-19 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
1KTV
 
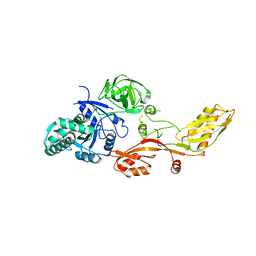 | |
1KV1
 
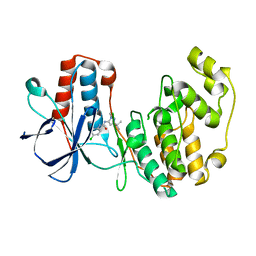 | | p38 MAP Kinase in Complex with Inhibitor 1 | | Descriptor: | 1-(5-TERT-BUTYL-2-METHYL-2H-PYRAZOL-3-YL)-3-(4-CHLORO-PHENYL)-UREA, p38 MAP kinase | | Authors: | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | Deposit date: | 2002-01-23 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
1KVI
 
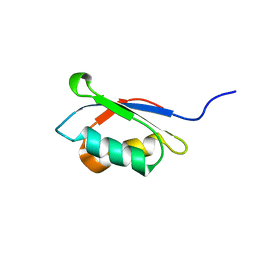 | |
1KW2
 
 | |
5IGR
 
 | | Macrolide 2'-phosphotransferase type I - complex with GDP and oleandomycin | | Descriptor: | (3S,5R,6S,7R,8R,11R,12S,13R,14S,15S)-6-HYDROXY-5,7,8,11,13,15-HEXAMETHYL-4,10-DIOXO-14-{[3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSYL]OXY}-1,9-DIOXASPIRO[2.13]HEXADEC-12-YL 2,6-DIDEOXY-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSIDE, AMMONIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2016-02-28 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Kinase-Mediated Macrolide Antibiotic Resistance.
Structure, 25, 2017
|
|
7S7F
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH DOT1L(998-1006) PHOSPHOPEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
5IH1
 
 | |
1KXL
 
 | |
1KXD
 
 | |
1KDE
 
 | | NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT, NMR, 22 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT | | Authors: | Sonnichsen, F.D, Deluca, C.I, Davies, P.L, Sykes, B.D. | | Deposit date: | 1996-07-08 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction.
Structure, 4, 1996
|
|
1KGF
 
 | |
1KX6
 
 | |
1KVC
 
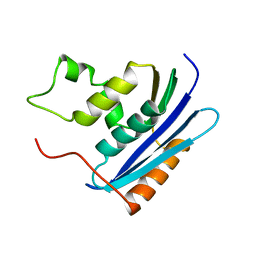 | | E. COLI RIBONUCLEASE HI D134N MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
7S7E
 
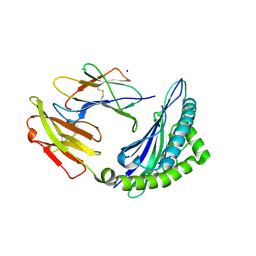 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH DOT1L(998-1006) PEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
1KAL
 
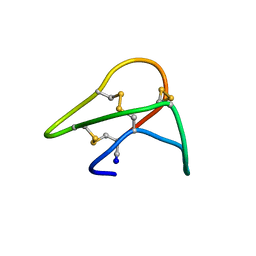 | |
1KGE
 
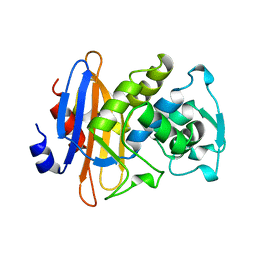 | |
1KVR
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
1KDF
 
 | | NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Sonnichsen, F.D, Deluca, C.I, Davies, P.L, Sykes, B.D. | | Deposit date: | 1996-07-08 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction.
Structure, 4, 1996
|
|
5ICF
 
 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine and sanguinarine | | Descriptor: | (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
5ICM
 
 | | 17beta-hydroxysteroid dehydrogenase type 14 in complex with a non-steroidal inhibitor | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bertoletti, N, Braun, F, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | New Insights into Human 17 beta-Hydroxysteroid Dehydrogenase Type 14: First Crystal Structures in Complex with a Steroidal Ligand and with a Potent Nonsteroidal Inhibitor.
J.Med.Chem., 59, 2016
|
|
1KXB
 
 | |
9FJU
 
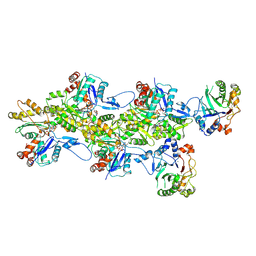 | | Structure of the DNase I- and phalloidin-bound pointed end of F-actin (conformer 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Boiero Sanders, M, Oosterheert, W, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Nat Commun, 15, 2024
|
|
1KVS
 
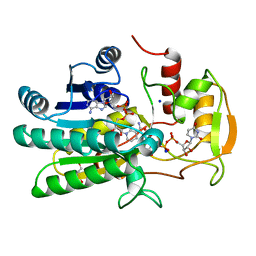 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
9FJY
 
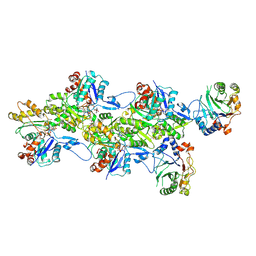 | | Structure of the DNase I- and phalloidin-bound pointed end of F-actin (conformer 2). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Boiero Sanders, M, Oosterheert, W, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Nat Commun, 15, 2024
|
|
