5ET7
 
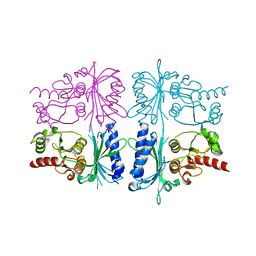 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8TUW
 
 | |
5FU2
 
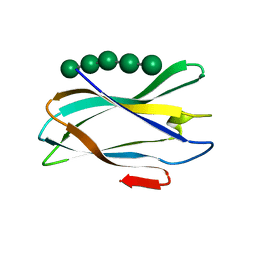 | | The complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition | | Descriptor: | CALCIUM ION, CBM74-RFGH5, SODIUM ION, ... | | Authors: | Basle, A, Luis, A.S, Venditto, I, Gilbert, H.J. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FWM
 
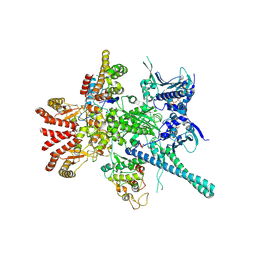 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
1RVI
 
 | | SOLUTION STRUCTURE OF THE DNA DODECAMER CGTTTTAAAACG | | Descriptor: | 5'-D(*CP*GP*TP*TP*TP*TP*AP*AP*AP*AP*CP*G)-3' | | Authors: | Stefl, R, Wu, H, Ravindranathan, S, Sklenar, V, Feigon, J. | | Deposit date: | 2003-12-13 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA A-tract bending in three dimensions: Solving the dA4T4 vs. dT4A4 conundrum.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6DLC
 
 | |
5CPQ
 
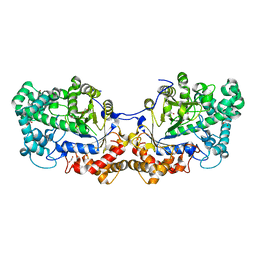 | | Disproportionating enzyme 1 from Arabidopsis - apo form | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
2AJD
 
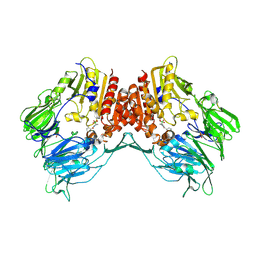 | | Porcine dipeptidyl peptidase IV (CD26) in complex with L-Pro-boro-L-Pro (boroPro) | | Descriptor: | (2R)-N-[(2R)-2-(DIHYDROXYBORYL)-1-L-PROLYLPYRROLIDIN-2-YL]-N-[(5R)-5-(DIHYDROXYBORYL)-1-L-PROLYLPYRROLIDIN-2-YL]-L-PROLINAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
6DJP
 
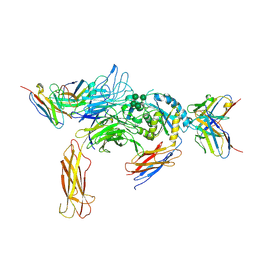 | | Integrin alpha-v beta-8 in complex with the Fabs 8B8 and 68 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 68 heavy chain Fab, ... | | Authors: | Cormier, A, Campbell, M.G, Nishimura, S.L, Cheng, Y. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of the alpha v beta 8 integrin reveals a mechanism for stabilizing integrin extension.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1YDR
 
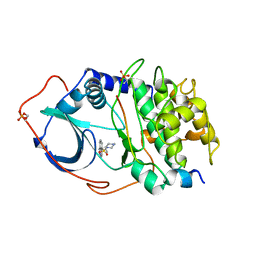 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H7 PROTEIN KINASE INHIBITOR 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE | | Descriptor: | 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE, C-AMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDT
 
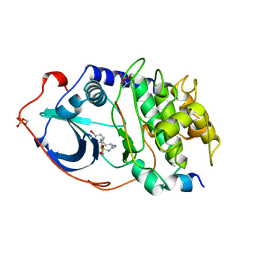 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H89 PROTEIN KINASE INHIBITOR N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDS
 
 | | Structure of CAMP-dependent protein kinase, alpha-catalytic subunit in complex with H8 protein kinase inhibitor [N-(2-methylamino)ethyl]-5-isoquinolinesulfonamide | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(METHYLAMINO)ETHYL]-5-ISOQUINOLINESULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
6DK0
 
 | | Human sigma-1 receptor bound to NE-100 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, GLYCEROL, N-{2-[4-methoxy-3-(2-phenylethoxy)phenyl]ethyl}-N-propylpropan-1-amine, ... | | Authors: | Schmidt, H.R, Kruse, A.C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for sigma1receptor ligand recognition.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1ZBI
 
 | | Bacillus halodurans RNase H catalytic domain mutant D132N in complex with 12-mer RNA/DNA hybrid | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3', 5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3', MAGNESIUM ION, ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
2CCW
 
 | | Crystal structure of Azurin II at atomic resolution (1.13 angstrom) | | Descriptor: | AZURIN II, COPPER (I) ION | | Authors: | Paraskevopoulos, K, Hough, M.A, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Active Site Structures and the Redox Properties of Blue Copper Proteins: Atomic Resolution Structure of Azurin II and Electronic Structure Calculations of Azurin, Plastocyanin and Stellacyanin.
Dalton Trans., 25, 2006
|
|
3NZM
 
 | |
1ZFV
 
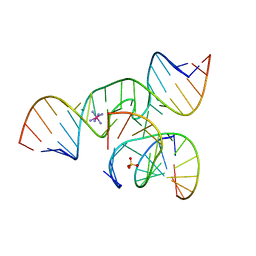 | | The structure of an all-RNA minimal Hairpin Ribozyme with Mutation G8A at the cleavage site | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*AP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Wedekind, J.E. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Water in the Active Site of an All-RNA Hairpin Ribozyme and Effects of Gua8 Base Variants on the Geometry of Phosphoryl Transfer.
Biochemistry, 45, 2006
|
|
6DWW
 
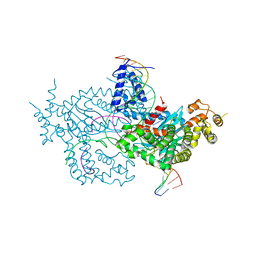 | | Hermes transposase deletion dimer complex with (A/T) DNA and Mn2+ | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3'), ... | | Authors: | Dyda, F, Voth, A.R, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
2C7I
 
 | |
6DX0
 
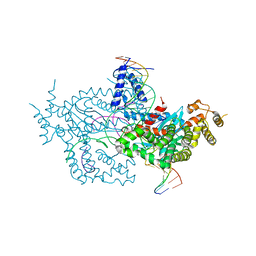 | | Hermes transposase deletion dimer complex with (A/T) DNA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3'), ... | | Authors: | Dyda, F, Voth, A.R, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
5IKR
 
 | | The Structure of Mefenamic Acid Bound to Human Cyclooxygenase-2 | | Descriptor: | 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Orlando, B.J, Malkowski, M.G. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Substrate-selective Inhibition of Cyclooxygeanse-2 by Fenamic Acid Derivatives Is Dependent on Peroxide Tone.
J.Biol.Chem., 291, 2016
|
|
1V7A
 
 | | Crystal structures of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-2-HYDROXY-1-[2-(2-NAPHTHYLOXY)ETHYL]PROPYL}-1H-IMIDAZONE-4-CARBOXAMIDE, ZINC ION, adenosine deaminase | | Authors: | Kinoshita, T. | | Deposit date: | 2003-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
6DJZ
 
 | | Human sigma-1 receptor bound to haloperidol | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, GLYCEROL, ... | | Authors: | Schmidt, H.R, Kruse, A.C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | Structural basis for sigma1receptor ligand recognition.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2CM5
 
 | | crystal structure of the C2B domain of rabphilin | | Descriptor: | CALCIUM ION, RABPHILIN-3A | | Authors: | Schlicker, C, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-05-04 | | Release date: | 2006-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The C2A-C2B Linker Defines the High Affinity Ca2+ Binding Mode of Rabphilin-3A.
J.Biol.Chem., 282, 2007
|
|
4CRX
 
 | |
